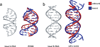Origins of specificity in protein-DNA recognition - PubMed (original) (raw)
Review
Origins of specificity in protein-DNA recognition
Remo Rohs et al. Annu Rev Biochem. 2010.
Abstract
Specific interactions between proteins and DNA are fundamental to many biological processes. In this review, we provide a revised view of protein-DNA interactions that emphasizes the importance of the three-dimensional structures of both macromolecules. We divide protein-DNA interactions into two categories: those when the protein recognizes the unique chemical signatures of the DNA bases (base readout) and those when the protein recognizes a sequence-dependent DNA shape (shape readout). We further divide base readout into those interactions that occur in the major groove from those that occur in the minor groove. Analogously, the readout of the DNA shape is subdivided into global shape recognition (for example, when the DNA helix exhibits an overall bend) and local shape recognition (for example, when a base pair step is kinked or a region of the minor groove is narrow). Based on the >1500 structures of protein-DNA complexes now available in the Protein Data Bank, we argue that individual DNA-binding proteins combine multiple readout mechanisms to achieve DNA-binding specificity. Specificity that distinguishes between families frequently involves base readout in the major groove, whereas shape readout is often exploited for higher resolution specificity, to distinguish between members within the same DNA-binding protein family.
Figures
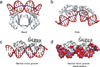
Figure 1. Molecular shape and electrostatic potential of A-DNA, B-DNA, and Z-DNA
The upper panels show the molecular shape in GRASP2 images (convex surfaces in green and concave surfaces in grey/black) [219] of the three helical forms of DNA constructed with 3DNA [91] from fiber diffraction data [71, 79]. Each DNA helix is comprised of 14-mers. The width and depth stated below were calculated with Curves [220, 221]. The lower panels show how the electrostatic potential at the molecular surface varies due to shape and atomic charges. The electrostatic potentials were calculated by solving the Poisson-Boltzmann equation with DelPhi [66, 222] at a salt concentration of 0.145 M (other parameters as described in Methods of [65]). Negative electrostatic potentials are shown in red and positive electrostatic potentials in blue. a. A-DNA with a narrow and deep major groove (2.2 Å wide; 9.5 Å deep) and a wide and shallow minor groove (10.9 Å wide; no defined depth). The model is of the alternating sequence d(GC)7. b. B-DNA (alternating sequence d(GC)7) with a wide and shallow major groove (5.9 Å wide; 5.5 Å deep) and a narrow and deep minor groove (11.4 Å wide; 4.0 Å deep). c. B-DNA (alternating sequence d(AT)7). Since the models are built based on fiber diffraction data, the shape of GC and AT alternating B-DNA does not reflect a sequence dependence. d. Z-DNA which lacks a major groove (13.2 Å wide; no defined depth), while the minor groove is narrow and deep (2.4 Å wide, 5.0 Å deep). The model is of the alternating sequence d(GC)7. e. A-DNA exhibits a strongly negative major groove but a hydrophobic minor groove surface, which is partially due to its exposed C3'-endo sugar moieties. f. B-DNA (alternating sequence d(GC)7) exhibits a negative minor groove and less negative major groove. g. B-DNA (alternating sequence d(AT)7). Variations in electrostatic potential between GC and AT alternating B-DNA reflect the different functional groups of the base pairs (e.g., positive guanine amino group in GC minor groove, neutral thymine methyl group in AT major groove). h. Z-DNA exhibits a negative minor groove and a positive surface on opposing edges of the bases.
Figure 2. Comparison of readout mechanisms based on local shape, bending, and kinking in protein-DNA complexes
a. HPV-18 E2 bound to DNA (PDB ID 1jj4) shows bending over a large stretch of the helix. The smooth curvature is visualized by the helix axis (blue) calculated with Curves [220]. b. The Lac repressor kinks the DNA at a central CpG base pair step stabilized by the partial intercalation of leucines (PDB ID 2kei). The helix axes calculated for both sides of the kink (blue) show an abrupt change in the helix trajectory caused by the kink. c. Phage 434 repressor recognizes local shape deformations of its operator with arginine residues (PDB ID 2or1) [65]. The narrow region of the minor groove that is contacted by arginines is highlighted in blue. d. For the same structure shown in c, the electrostatic potential of the operator calculated in the absence of the repressor is plotted on the molecular surface. In comparison with Figures 1f and 1g, the bottom of the minor groove is only red indicating enhanced negative electrostatic potential [65].
Figure 3. DNAs bound to proteins have features already present in unbound DNAs
a. The structure of the unbound FIN-B sequence (PDB ID 2b1c) is similar to ideal A-DNA (grey), while the bound structure of the Zif286-DNA complex (PDB ID 1a1f) has some A-DNA characteristics, notably a wider minor groove than normally found in B-DNA. b. The specific HPV-18 E2 site (PDB ID 1ilc) contains an A-tract AATT in the central region of the helix, which, although not contacted by the protein, bends the free-DNA structure (red) in a similar manner as seen in the bound structure (blue) of the HPV-18 E2-DNA complex (PDB ID 1jj4). In comparison to ideal B-DNA (grey), the bending is reflected by a minor groove narrowing in the center of the free and bound DNA.
Figure 4. Base recognition in the major and minor groove
Sequence specific patterns on the edges of the bases in the major groove underlie the ability for proteins to readout base pairs through hydrogen bonds and hydrophobic contacts (hydrogen bond acceptors in red, donors in blue, thymine methyl group in yellow, and base carbon hydrogens in white). In contrast, A:T versus T:A and C:G versus G:C are indistinguishable in the minor groove. The three panels show successive rotations of 90° around the helix axis. The dodecamer d(GACT)3 was built based on fiber diffraction data with 3DNA [91].
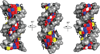
Figure 5. Hox DNA binding specificity mediated by local shape recognition
All panels show either the fkh250 binding site or the fkh250con binding site. fkh250, but not fkh250con, has two minor groove minima, which creates a more negative electrostatic potential (minus signs). “W” refers to the Hox YPWM motif that makes a direct contact with the cofactor Exd. See [29] for details. a. In the absence of Exd, Scr does not bind with high affinity to fkh250 because the arginines on the N-terminal arm and linker of Scr are not positioned correctly. b. Other Hox proteins do not bind well to fkh250 even in the presence of Exd because their Nterminal arms and linker regions do not have the correct residues. c. The Scr/Exd heterodimer binds well to fkh250 because the Scr N-terminal arm and linker region have the correct residues, and Exd positions them correctly by binding the YPWM motif (W). d. Other Hox/Exd heterodimers bind well to fkh250con. This binding site is not as selective because it has a less negative electrostatic potential. Thus, the sequences of the Hox N-terminal arms and linker regions are not as important for binding.
Figure 5. Hox DNA binding specificity mediated by local shape recognition
All panels show either the fkh250 binding site or the fkh250con binding site. fkh250, but not fkh250con, has two minor groove minima, which creates a more negative electrostatic potential (minus signs). “W” refers to the Hox YPWM motif that makes a direct contact with the cofactor Exd. See [29] for details. a. In the absence of Exd, Scr does not bind with high affinity to fkh250 because the arginines on the N-terminal arm and linker of Scr are not positioned correctly. b. Other Hox proteins do not bind well to fkh250 even in the presence of Exd because their Nterminal arms and linker regions do not have the correct residues. c. The Scr/Exd heterodimer binds well to fkh250 because the Scr N-terminal arm and linker region have the correct residues, and Exd positions them correctly by binding the YPWM motif (W). d. Other Hox/Exd heterodimers bind well to fkh250con. This binding site is not as selective because it has a less negative electrostatic potential. Thus, the sequences of the Hox N-terminal arms and linker regions are not as important for binding.
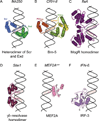
Figure 6. Examples of minor groove shape recognition
Each panel shows a different example in which basic residues bind to minor grooves. a. Arginine residues present on Scr’s N-terminal arm and linker region requires heterodimerzation with Exd to be positioned correctly to insert into a narrow minor groove region of fkh250. (PDB ID 2r5z) b. Arginine residues present on the linker region that separates POUHD from POUS of Brn-5 insert into a narrow minor groove of the CRH-II binding site. (PDB ID 3d1n) c. Arginine residues present on a C-terminal extension of a MogR homodimer insert into narrow regions of the flaA binding site. (PDB ID 3fdq) d. An N-terminal extension from the γδ resolvase has an arginine that inserts into a narrow minor groove and a second arginine that inserts into the major groove of its binding site. (PDB ID 1gdt) e. MEF2A recognizes a narrow minor groove of the MEF2A binding site via an arginine and glycine present on an N-terminal strand and via a lysine present on alpha helix α1. (PDB ID 1egw) f. A histidine residue of IRF-3 inserts into a narrow minor groove region of the IFN-β enhancer. (PDB ID 1t2k)
Similar articles
- P22 c2 repressor-operator complex: mechanisms of direct and indirect readout.
Watkins D, Hsiao C, Woods KK, Koudelka GB, Williams LD. Watkins D, et al. Biochemistry. 2008 Feb 26;47(8):2325-38. doi: 10.1021/bi701826f. Epub 2008 Feb 1. Biochemistry. 2008. PMID: 18237194 - Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: DNA binding specificity based on energetics of DNA kinking.
Chen S, Vojtechovsky J, Parkinson GN, Ebright RH, Berman HM. Chen S, et al. J Mol Biol. 2001 Nov 16;314(1):63-74. doi: 10.1006/jmbi.2001.5089. J Mol Biol. 2001. PMID: 11724532 - The role of DNA shape in protein-DNA recognition.
Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. Rohs R, et al. Nature. 2009 Oct 29;461(7268):1248-53. doi: 10.1038/nature08473. Nature. 2009. PMID: 19865164 Free PMC article. - DNA curving and bending in protein-DNA recognition.
Harrington RE. Harrington RE. Mol Microbiol. 1992 Sep;6(18):2549-55. doi: 10.1111/j.1365-2958.1992.tb01431.x. Mol Microbiol. 1992. PMID: 1333034 Review. - Making the bend: DNA tertiary structure and protein-DNA interactions.
Harteis S, Schneider S. Harteis S, et al. Int J Mol Sci. 2014 Jul 14;15(7):12335-63. doi: 10.3390/ijms150712335. Int J Mol Sci. 2014. PMID: 25026169 Free PMC article. Review.
Cited by
- DNA-Directed Protein Packing within Single Crystals.
Winegar PH, Hayes OG, McMillan JR, Figg CA, Focia PJ, Mirkin CA. Winegar PH, et al. Chem. 2020 Apr 9;6(4):1007-1017. doi: 10.1016/j.chempr.2020.03.002. Epub 2020 Mar 23. Chem. 2020. PMID: 33709040 Free PMC article. - Protein-DNA docking with a coarse-grained force field.
Setny P, Bahadur RP, Zacharias M. Setny P, et al. BMC Bioinformatics. 2012 Sep 11;13:228. doi: 10.1186/1471-2105-13-228. BMC Bioinformatics. 2012. PMID: 22966980 Free PMC article. - Statistical analysis of structural determinants for protein-DNA-binding specificity.
Corona RI, Guo JT. Corona RI, et al. Proteins. 2016 Aug;84(8):1147-61. doi: 10.1002/prot.25061. Epub 2016 Jun 15. Proteins. 2016. PMID: 27147539 Free PMC article. - Initial DNA interactions of the binuclear threading intercalator Λ,Λ-[μ-bidppz(bipy)4Ru2]4+: an NMR study with [d(CGCGAATTCGCG)]2.
Wu L, Reymer A, Persson C, Kazimierczuk K, Brown T, Lincoln P, Nordén B, Billeter M. Wu L, et al. Chemistry. 2013 Apr 22;19(17):5401-10. doi: 10.1002/chem.201203175. Epub 2013 Feb 28. Chemistry. 2013. PMID: 23447081 Free PMC article. - Transcriptional reprogramming by oxidative stress occurs within a predefined chromatin accessibility landscape.
Levings DC, Lacher SE, Palacios-Moreno J, Slattery M. Levings DC, et al. Free Radic Biol Med. 2021 Aug 1;171:319-331. doi: 10.1016/j.freeradbiomed.2021.05.016. Epub 2021 May 13. Free Radic Biol Med. 2021. PMID: 33992677 Free PMC article.
References
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054510-19/GM/NIGMS NIH HHS/United States
- R01 GM054510/GM/NIGMS NIH HHS/United States
- GM54510/GM/NIGMS NIH HHS/United States
- R01 GM054510-17/GM/NIGMS NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM030518/GM/NIGMS NIH HHS/United States
- U54 CA121852-05/CA/NCI NIH HHS/United States
- R01 GM030518-29/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources