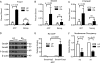Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells - PubMed (original) (raw)
Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells
Joseph H Taube et al. J Biol Chem. 2010.
Abstract
Epigenetic control of genes that are silent in embryonic stem cells, but destined for expression during differentiation, includes distinctive hallmarks, such as simultaneous activating/repressing (bivalent) modifications of chromatin and DNA hypomethylation at enhancers of gene expression. Although alpha-fetoprotein (Afp) falls into this class of genes, as it is silent in pluripotent stem cells and activated during differentiation of endoderm, we find that Afp chromatin lacks bivalent histone modifications. However, critical regulatory sites for Afp activation, overlapping Foxa1/p53/Smad-binding elements, are located within a 300-bp region lacking DNA methylation, due to transposed elements underrepresented in CpG sequences: a short interspersed transposable element and a medium reiterated sequence 1 element. Forkhead family member Foxa1 is activated by retinoic acid treatment of embryonic stem cells, binds its DNA consensus site within the short interspersed transposable/medium reiterated sequence 1 elements, and displaces linker histone H1 from silent Afp chromatin. Small interfering RNA depletion of Foxa1 showed that Foxa1 is essential in providing chromatin access to transforming growth factor beta-activated Smad2 and Smad4 and their subsequent DNA binding. Together these transcription factors establish highly acetylated chromatin and promote expression of Afp. Foxa1 acts as a pioneer transcription factor in de novo activation of Afp, by exploiting a lack of methylation at juxtaposed transposed elements, to bind and poise chromatin for intersection with transforming growth factor beta signaling during differentiation of embryonic stem cells.
Figures
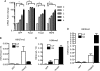
FIGURE 1.
ES cells express Afp when treated with RA and chromatin at the distal promoter is not methylated at H3K4, Lys-9, or Lys-27. A, RT-PCR was performed on cells treated with RA for 0, 1, 2, 3, or 4 days. Ct (threshold cycle) values for each of the indicated genes were normalized to 18S. * indicates significant difference from 0 days of RA treatment. B–D, chromatin immunoprecipitation was performed on ES cells maintained in LIF or treated with RA for 4 days. DNA from immunoprecipitations for H3K27me2 (B), H3K4me3 (C), and H3K9me2 (D) was analyzed by real time PCR for amplification of the Afp distal promoter with the Hoxa3 and Trappc9 promoters as positive controls. Levels of each modification are expressed as a ratio to levels of histone H3, determined by a separate immunoprecipitation. The y axis in D is split to better indicate the low values in Afp samples. Error bars represent S.D. of at least three repetitions.
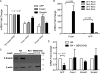
FIGURE 2.
Repetitive, transposable elements define a methylation-free Foxa1 regulatory unit. A, DNA methylation analysis of the Afp distal promoter. A diagram of the Afp promoter is shown on the top, with locations of the Foxa1-binding site (light gray bar, approximately −850) and transcription start site (arrow, +1) shown. Each vertical line represents a single CpG site. The dark gray bar indicates the region analyzed by bisulfite sequencing. Three sets of bisulfite-sequencing results are shown with each row representing an individual cloned allele. Circles represent CpG sites and their spacing accurately reflects the CpG density of the region: black circle, methylated CpG site; white circle, unmethylated CpG site. The percentage of methylated alleles at each CpG is shown at the bottom. Each set of DNA methylation analyses shown was performed in ES cells maintained in LIF (top), in cells treated with RA for 2 days (middle), and cells treated with RA for 4 days (bottom). B, the Foxa regulatory element centered at −850 in the Afp promoter overlaps with consensus Smad binding elements and p53 response elements. Alignment with the transposable SINE and MER1 elements is shown at the bottom, corresponding to the CpG-free region from −1089 to −793 of the Afp distal promoter. C, ChIP was performed on ES cells maintained in LIF or treated with RA for 4 days. DNA from ChIP of Foxd3 was analyzed by real time PCR for binding to the Afp distal promoter and the ALB promoter and was graphed as percent bound minus IgG relative to input. Error bars represent S.D. of at least three repetitions.
FIGURE 3.
Foxa1, Smad4, and TGF-β signaling are required for activation of Afp expression. A, RT-PCR was performed on RNA extracted from RA-treated ES cells transfected with Foxa1-targeted siRNA oligos, Smad4-target siRNA oligos, or scrambled siRNA oligos. Ct values from amplification of the indicated genes were normalized to 18S and graphed relative to the non-target siRNA. Error bars represent standard deviation from at least three repetitions. B, RT-PCR was performed on RNA extracted from Foxa1 null and wild type ES cells treated with RA for 0 or 4 days. Ct values from amplification of the indicated genes were normalized to 18S and graphed relative to the 0 day wild type sample. C, Western blots for P-Smad2 and β-actin were performed on protein lysates from ES cells treated with RA ± SB431542 for 4 days. D, RT-PCR was performed on RNA extracted from ES cells treated with RA ± SB431542 for 4 days. Expression of the indicated genes was normalized to 18S and graphed relative to the RA-treated sample.
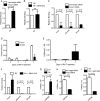
FIGURE 4.
Activated Smad proteins bind chromatin at the Afp distal promoter only when Foxa1 is bound and silent chromatin is altered. A–C, E, and F, chromatin immunoprecipitation was performed on ES cells maintained in LIF or treated with RA for 4 days. DNA from immunoprecipitations for Foxa1 (A), Smad4 (B), P-Smad2 (C), H3 and H1 (F) was analyzed by real time PCR for binding to the Afp distal promoter and to the Nanog p53 response element. Results were graphed as percent bound minus IgG relative to input (A-C) or as normalized to LIF (F). E, re-ChIP analysis reveals simultaneous association of Foxa1 and Smad4 at the AFP distal promoter in RA-treated ES cells, but not in LIF-treated ES cells. A 1° ChIP was done with Smad4 and reactions were pooled, released from protein A beads, and then reimmunoprecipitated for IgG or Foxa1. Error bars represent S.D. of at least three repetitions; ns, not significant. D, whole cell lysates from ES cells maintained with LIF or differentiated with RA were probed with antibodies to Smad4, Smad2, P-Smad2, and β-actin. Triangles indicate differential loading.
FIGURE 5.
Foxa1 acts upstream of Smads to mediate Smad binding, nucleosome occupancy reduction, and H3K9 acetylation. Chromatin immunoprecipitation was performed on ES cells treated with RA for 4 days and transfected with either Foxa1-targeted or scrambled siRNA oligos (A, C, and H) or treated with SB431542 or dimethyl sulfoxide (B, F, and I). Transfection of the siRNA oligos for Foxa1 or for a non-target control and treatment with SB431542 or dimethyl sulfoxide occurred coincident with addition of RA. DNA from immunoprecipitations for H3 (A and B), Foxa1, P-Smad2, and Smad4 (C, D, and F), and H3 and H3K9ac (E and G–I) was analyzed by real time PCR for binding to the Afp distal promoter. DNA from immunoprecipitations was analyzed by real time PCR for binding to the Afp distal promoter. Levels of acH3K9 are expressed as a ratio to levels of histone H3, determined by a separate immunoprecipitation. Error bars represent S.D. from at least three repetitions. ns, not significant.
Similar articles
- A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene.
Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC. Wilkinson DS, et al. Mol Cell Biol. 2005 Feb;25(3):1200-12. doi: 10.1128/MCB.25.3.1200-1212.2005. Mol Cell Biol. 2005. PMID: 15657445 Free PMC article. - Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers.
Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, Salbert G, Eeckhoute J. Sérandour AA, et al. Genome Res. 2011 Apr;21(4):555-65. doi: 10.1101/gr.111534.110. Epub 2011 Jan 13. Genome Res. 2011. PMID: 21233399 Free PMC article. - Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-beta-mediated transcription repression.
Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Wilkinson DS, et al. Mol Cell Biol. 2008 Mar;28(6):1988-98. doi: 10.1128/MCB.01442-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212064 Free PMC article. - TGF-β control of stem cell differentiation genes.
Massagué J, Xi Q. Massagué J, et al. FEBS Lett. 2012 Jul 4;586(14):1953-8. doi: 10.1016/j.febslet.2012.03.023. Epub 2012 Apr 10. FEBS Lett. 2012. PMID: 22710171 Free PMC article. Review. - Pioneer factors in embryonic stem cells and differentiation.
Smale ST. Smale ST. Curr Opin Genet Dev. 2010 Oct;20(5):519-26. doi: 10.1016/j.gde.2010.06.010. Epub 2010 Jul 16. Curr Opin Genet Dev. 2010. PMID: 20638836 Free PMC article. Review.
Cited by
- Multimodal epigenetic changes and altered NEUROD1 chromatin binding in the mouse hippocampus underlie FOXG1 syndrome.
Akol I, Izzo A, Gather F, Strack S, Heidrich S, Ó hAilín D, Villarreal A, Hacker C, Rauleac T, Bella C, Fischer A, Manke T, Vogel T. Akol I, et al. Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2122467120. doi: 10.1073/pnas.2122467120. Epub 2023 Jan 4. Proc Natl Acad Sci U S A. 2023. PMID: 36598943 Free PMC article. - Transposable Elements in Human Cancer: Causes and Consequences of Deregulation.
Anwar SL, Wulaningsih W, Lehmann U. Anwar SL, et al. Int J Mol Sci. 2017 May 4;18(5):974. doi: 10.3390/ijms18050974. Int J Mol Sci. 2017. PMID: 28471386 Free PMC article. Review. - Different subpopulations of regulatory T cells in human autoimmune disease, transplantation, and tumor immunity.
Jiang Z, Zhu H, Wang P, Que W, Zhong L, Li XK, Du F. Jiang Z, et al. MedComm (2020). 2022 Apr 21;3(2):e137. doi: 10.1002/mco2.137. eCollection 2022 Jun. MedComm (2020). 2022. PMID: 35474948 Free PMC article. Review. - Significance of the NOR1-FOXA1/HDAC2-Slug regulatory network in epithelial-mesenchymal transition of tumor cells.
Wang W, Yi M, Chen S, Li J, Li G, Yang J, Zheng P, Zhang H, Xiong W, McCarthy JB, Li G, Li X, Xiang B. Wang W, et al. Oncotarget. 2016 Mar 29;7(13):16745-59. doi: 10.18632/oncotarget.7778. Oncotarget. 2016. PMID: 26934447 Free PMC article. - The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation.
Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. Iwafuchi-Doi M, et al. Mol Cell. 2016 Apr 7;62(1):79-91. doi: 10.1016/j.molcel.2016.03.001. Mol Cell. 2016. PMID: 27058788 Free PMC article.
References
- Keohane A. M., O'Neill L. P., Belyaev N. D., Lavender J. S., Turner B. M. (1996) Dev. Biol. 180, 618–630 - PubMed
- Lee J. H., Hart S. R., Skalnik D. G. (2004) Genesis 38, 32–38 - PubMed
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 - PubMed
- Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Cell 125, 301–313 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM081627/GM/NIGMS NIH HHS/United States
- T32 HD007325/HD/NICHD NIH HHS/United States
- P01 GM081627/GM/NIGMS NIH HHS/United States
- R01 GM053683/GM/NIGMS NIH HHS/United States
- GM53683/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous