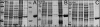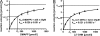A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyl-transferase reaction - PubMed (original) (raw)
A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyl-transferase reaction
Elisa Haug-Schifferdecker et al. J Biol Chem. 2010.
Abstract
Five fungal genomes from the Ascomycota (sac fungi) were found to contain a gene with sequence similarity to a recently discovered small group of bacterial prenyltransferases that catalyze the C-prenylation of aromatic substrates in secondary metabolism. The genes from Aspergillus terreus NIH2624, Botryotinia fuckeliana B05.10 and Sclerotinia sclerotiorum 1980 were expressed in Escherichia coli, and the resulting His(8)-tagged proteins were purified and investigated biochemically. Their substrate specificity was found to be different from that of any other prenyltransferase investigated previously. Using 2,7-dihydroxynaphthalene (2,7-DHN) and dimethylallyl diphosphate as substrates, they catalyzed a regiospecific Friedel-Crafts alkylation of 2,7-DHN at position 3. Using the enzyme of A. terreus, the K(m) values for 2,7-DHN and dimethylallyl diphosphate were determined as 324 +/- 25 microM and 325 +/- 35 microM, respectively, and k(cat) as 0.026 +/- 0.001 s(-1). A significantly lower level of prenylation activity was found using dihydrophenazine-1-carboxylic acid as aromatic substrate, and only traces of products were detected with aspulvinone E, flaviolin, or 4-hydroxybenzoic acid. No product was formed with l-tryptophan, l-tyrosine, or 4-hydroxyphenylpyruvate. The genes for these fungal prenyltransferases are not located within recognizable secondary metabolic gene clusters. Their physiological function is yet unknown.
Figures
FIGURE 1.
Reactions catalyzed by the bacterial enzymes CloQ, NphB, and SCO7190 and by the fungal aspulvinone dimethylallyltransferase. The genuine aromatic substrate of NphB in the biosynthesis of naphterpin is unknown; 2,7-dihydroxynaphthalene is one of the artificial substrates accepted in vitro. GPP, geranyl diphosphate.
FIGURE 2.
A, sequence comparison of the bacterial prenyltransferases NphB, CloQ, and NovQ with the fungal proteins PtfAt, PtfBf, PtfSs, PtfPm, and PtfMc. For PtfAt, both the current sequence in the database (PtfAt-Or) and the corrected sequence (PtfAt) are displayed. The amino acids of NphB involved in the binding of the pyrophosphate moiety (▴, Lys119, Asn173, Tyr216, Arg228, and Lys284; over H2O ■, Tyr282) and in the coordination of the Mg2+ ion (●, Asp62) are marked. Likewise, the position of the lysine residue of CloQ and NovQ, suggested to functionally replace the positive charge of the Mg2+ ion, is marked (★). The sequence was created with Clustal W2 (22) and visualized by ESPript 2.2 (23). The sequence shows the secondary structure elements of NphB: α, α helices; η, 310 helices; β, β strands; TT, strict β turns. Strict sequence identity is shown by a black box with a white character, and similarity is shown by bold characters in a black frame. B, sequence identities (%) of the predicted gene products of the fungal genes _ptf_At, _ptf_Bf, _ptf_Ss, _ptf_Pm-S, and _ptf_Mc with the bacterial ABBA prenyltransferases NphB, CloQ, and NovQ. For _ptf_Pm, an N-terminally truncated version (_ptf_Pm-S) was used for this calculation (see text).
FIGURE 3.
Purification of the fungal prenyltransferases PtfAt (A), PtfBf (B), and PtfSs (C) after expression in the form of His8-tagged fusion proteins. Lane 1, molecular mass standards; lane 2, total protein before IPTG induction; lane 3, total protein after IPTG induction; lane 4, soluble protein after IPTG induction; lane 5, insoluble protein after IPTG induction; lane 6, protein after Ni2+ affinity chromatography; lane 7, molecular mass standards; lane 8, protein after ion exchange chromatography. The calculated masses are 36.6 kDa for PtfAt, 36.7 kDa for PtfSs, and 36.5 kDa for PtfBf. The 12% polyacrylamide gel was stained with Coomassie Brilliant Blue R-250.
FIGURE 4.
A, the aromatic prenylation reaction catalyzed by PtfAt. B, HPLC analysis of prenyltransferase assays with PtfAt. Detection: UV, 285 nm.
FIGURE 5.
Determination of Km values of PtfAt for DMAPP and 2,7-dihydroxynaphthalene. In A, 2,7-diydroxynaphthalene was kept constant at 2 m
m
. In B, DMAPP was kept constant at 2 m
m
. Km and _k_cat values were determined by nonlinear regression, using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA).
Similar articles
- The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria.
Metzger U, Schall C, Zocher G, Unsöld I, Stec E, Li SM, Heide L, Stehle T. Metzger U, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14309-14. doi: 10.1073/pnas.0904897106. Epub 2009 Aug 12. Proc Natl Acad Sci U S A. 2009. PMID: 19706516 Free PMC article. - Biochemical characterization of indole prenyltransferases: filling the last gap of prenylation positions by a 5-dimethylallyltryptophan synthase from Aspergillus clavatus.
Yu X, Liu Y, Xie X, Zheng XD, Li SM. Yu X, et al. J Biol Chem. 2012 Jan 6;287(2):1371-80. doi: 10.1074/jbc.M111.317982. Epub 2011 Nov 28. J Biol Chem. 2012. PMID: 22123822 Free PMC article. - CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterisation.
Yin WB, Ruan HL, Westrich L, Grundmann A, Li SM. Yin WB, et al. Chembiochem. 2007 Jul 9;8(10):1154-61. doi: 10.1002/cbic.200700079. Chembiochem. 2007. PMID: 17525915 - Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives.
Steffan N, Grundmann A, Yin WB, Kremer A, Li SM. Steffan N, et al. Curr Med Chem. 2009;16(2):218-31. doi: 10.2174/092986709787002772. Curr Med Chem. 2009. PMID: 19149573 Review. - Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives.
Li SM. Li SM. Phytochemistry. 2009 Oct-Nov;70(15-16):1746-57. doi: 10.1016/j.phytochem.2009.03.019. Epub 2009 May 3. Phytochemistry. 2009. PMID: 19398116 Review.
Cited by
- A basidomycetous hydroxynaphthalene-prenylating enzyme exhibits promiscuity toward prenyl donors.
Martin A, Dierlamm N, Zocher G, Li SM. Martin A, et al. Appl Microbiol Biotechnol. 2023 Aug;107(15):4845-4852. doi: 10.1007/s00253-023-12621-1. Epub 2023 Jun 16. Appl Microbiol Biotechnol. 2023. PMID: 37326682 Free PMC article. - Synthetic biology, combinatorial biosynthesis, and chemo‑enzymatic synthesis of isoprenoids.
Malico AA, Calzini MA, Gayen AK, Williams GJ. Malico AA, et al. J Ind Microbiol Biotechnol. 2020 Oct;47(9-10):675-702. doi: 10.1007/s10295-020-02306-3. Epub 2020 Sep 3. J Ind Microbiol Biotechnol. 2020. PMID: 32880770 Free PMC article. Review. - Enzymatic studies on aromatic prenyltransferases.
Mori T. Mori T. J Nat Med. 2020 Jun;74(3):501-512. doi: 10.1007/s11418-020-01393-x. Epub 2020 Mar 17. J Nat Med. 2020. PMID: 32180104 Free PMC article. Review. - Vib-PT, an Aromatic Prenyltransferase Involved in the Biosynthesis of Vibralactone from Stereum vibrans.
Bai N, Li GH, Luo SL, Du L, Hu QY, Xu HK, Zhang KQ, Zhao PJ. Bai N, et al. Appl Environ Microbiol. 2020 May 5;86(10):e02687-19. doi: 10.1128/AEM.02687-19. Print 2020 May 5. Appl Environ Microbiol. 2020. PMID: 32144102 Free PMC article. - Comparative proteomic analysis reveals the regulatory network of the veA gene during asexual and sexual spore development of Aspergillus cristatus.
Liu H, Sang S, Wang H, Ren X, Tan Y, Chen W, Liu Z, Liu Y. Liu H, et al. Biosci Rep. 2018 Jul 31;38(4):BSR20180067. doi: 10.1042/BSR20180067. Print 2018 Aug 31. Biosci Rep. 2018. PMID: 29773679 Free PMC article. Retracted.
References
- Saleh O., Haagen Y., Seeger K., Heide L. (2009) Phytochemistry 70, 1728–1738 - PubMed
- Heide L. (2009) Curr. Opin. Chem. Biol. 13, 171–179 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous