Human population differentiation is strongly correlated with local recombination rate - PubMed (original) (raw)
Human population differentiation is strongly correlated with local recombination rate
Alon Keinan et al. PLoS Genet. 2010.
Abstract
Allele frequency differences across populations can provide valuable information both for studying population structure and for identifying loci that have been targets of natural selection. Here, we examine the relationship between recombination rate and population differentiation in humans by analyzing two uniformly-ascertained, whole-genome data sets. We find that population differentiation as assessed by inter-continental F(ST) shows negative correlation with recombination rate, with F(ST) reduced by 10% in the tenth of the genome with the highest recombination rate compared with the tenth of the genome with the lowest recombination rate (P<<10(-12)). This pattern cannot be explained by the mutagenic properties of recombination and instead must reflect the impact of selection in the last 100,000 years since human continental populations split. The correlation between recombination rate and F(ST) has a qualitatively different relationship for F(ST) between African and non-African populations and for F(ST) between European and East Asian populations, suggesting varying levels or types of selection in different epochs of human history.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
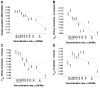
Figure 1. Population differentiation in allele frequencies is inversely correlated with recombination rate.
We placed 1,110,338 SNPs into 10 bins according to the recombination rate in a 3 Mb window centered on each SNP. The x-axis of all panels indicates the recombination rate, with the values indicated on the ticks corresponding to the edges between 10 bins. For each bin, at an x-axis position corresponding to the median recombination rate across the SNPs at that bin, the figure presents (A) global population differentiation between African Americans, Europeans, and Chinese; (B) F ST between African Americans and Europeans; (C) F ST between African Americans and Chinese; and (D) F ST between Europeans and Chinese. Error bars indicate ±1 standard error, which is estimated based on 1,000 moving block bootstraps over the SNPs in the bin. Linear regression of F ST estimates as a function of median recombination rate in each bin is also presented (solid line) and corresponds to (A) 0.1280–0.0048ρ (B) 0.1138–0.0057ρ (C) 0.1546–0.0067ρ and (D) 0.1156–0.0022ρ. The corresponding correlation coefficient estimates between F ST and median recombination rate are (A) r = −0.962 (P = 8.9×10−6), (B) −0.815 (P = 0.0041), (C) −0.931 (P = 0.0001), and (D) −0.361 (P = 0.306). For comparison, population differentiation based on all SNPs in all bins combined is also presented (horizontal dotted line). The y-axis range is different between the four panels but spans 0.02 units in all. Figure S1 repeats Figure 1A for sets of SNPs of different minor allele frequency categories.
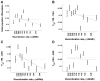
Figure 2. Population differentiation is more strongly correlated with recombination rate in genes than outside of genes.
Global population differentiation between African Americans, Europeans, and Chinese is presented for coding SNPs (cSNPs). Except for focusing on the 21,391 SNPs in coding exons, the figure is identical to Figure 1A. In addition to the linear regression of F ST estimates as a function of the median recombination rate in each bin (solid line; 0.1381–0.0081ρ), the linear regression for the rest of the data set (non-coding SNPs) is provided (dashed line; 0.1278–0.0048ρ), which is very similar to the regression based on the entire data set (Figure 1A). The correlation coefficient between F ST of cSNPs and median recombination rate is −0.752 (P = 0.012).
Figure 3. Confirmation of the correlation of allele frequency differentiation and recombination rate in uniformly-ascertained subsets of HapMap.
Similar to Figure 1, we placed 248,886 uniformly-ascertained HapMap SNPs into 10 bins according to the recombination rate and estimated for each bin (A) global population differentiation between YRI, CEU, and ASN (ASN denotes the combined CHB and JPT samples); (B) F ST between YRI and CEU; (C) F ST between YRI and ASN; and (D) F ST between CEU and ASN. Linear regression as a function of the median recombination rate (solid line) is (A) 0.1473–0.0026ρ (B) 0.1541–0.0028ρ (C) 0.1819–0.0046ρ, and (D) 0.1060–0.0005ρ. The corresponding correlation coefficient estimate between F ST and median recombination rate is (A) r = −0.526 (P = 0.118), (B) −0.482 (P = 0.158), (C) −0.634 (P = 0.049), and (D) −0.066 (P = 0.857).
Figure 4. The relationship between population differentiation and recombination rate in the larger set of HapMap 3 populations.
We placed 1,326,404 autosomal HapMap 3 SNPs (release 2) into 10 bins according to recombination rate and estimated for each bin (A) global population differentiation between all 11 populations, and (B–E) population differentiation between pairs of populations. To avoid clutter, (B–E) depict only the linear (dashed lines) and quadratic (solid lines) regression of F ST estimates as a function of the median recombination rate in each bin and partition the populations-pairs as follows: (B) F ST between an African and a non-African population, where a negative correlation is observed with recombination rate, and where the quadratic regression is convex, (C) F ST between a population of European, East Asian, or South Asian ancestry and a second population of a different one of these three ancestries, which shows a concave quadratic regression for all pairs of populations, and which recapitulates the result observed between North Europeans and East Asians in the uniformly-ascertained datasets (Figure 1D and Figure 3D). (A weaker phenomenon is observed for the South Asian GIH sample, which may be due to this population being somewhat related to both Europeans and East Asians , thereby confounding the North European–East Asian signal), (D) F ST between two African populations, which shows a much steeper linear regression compared to intercontinental F ST, as well as a convex quadratic regression, and (E) F ST between closely-related non-African populations (within either Europe or East Asia; genome-wide F ST<0.008), showing a very steep linear regression and a convex quadratic regression. F ST based on all SNPs in all bins combined is presented as a horizontal dotted line and is equal to 1 in panels B–E since these present normalized F ST values obtained by dividing each value by the genome-wide F ST for the same pair of populations. Population codes are as follows: WAF (“West African”) is a combined sample of YRI (Yoruba in Ibadan, Nigeria) and LWK (Luhya in Webuye, Kenya); EAS (“East Asia”) is a combined sample of CHB (Han Chinese in Beijing, China), CHD (Chinese in Metropolitan Denver, CO, USA), and JPT (Japanese in Tokyo, Japan); EUR (“Europe”) is a combined sample of CEU (ancestry from Northern and Western Europe) and TSI (Toscani in Italia); GIH is a sample of Gujarati Indians in Houston, TX, USA; MKK is a sample of Maasai in Kinyawa, Kenya; and CHI (Chinese) is a combined sample of CHB and CHD.
Similar articles
- Population-specific common SNPs reflect demographic histories and highlight regions of genomic plasticity with functional relevance.
Choudhury A, Hazelhurst S, Meintjes A, Achinike-Oduaran O, Aron S, Gamieldien J, Jalali Sefid Dashti M, Mulder N, Tiffin N, Ramsay M. Choudhury A, et al. BMC Genomics. 2014 Jun 6;15(1):437. doi: 10.1186/1471-2164-15-437. BMC Genomics. 2014. PMID: 24906912 Free PMC article. - Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.
Pimenoff VN, Laval G, Comas D, Palo JU, Gut I, Cann H, Excoffier L, Sajantila A. Pimenoff VN, et al. Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684 - Genetic Structural Differentiation Analyses of Intercontinental Populations and Ancestry Inference of the Chinese Hui Group Based on a Novel Developed Autosomal AIM-InDel Genotyping System.
Xie T, Shen C, Jin X, Lan Q, Fang Y, Zhu B. Xie T, et al. Biomed Res Int. 2020 Aug 25;2020:2124370. doi: 10.1155/2020/2124370. eCollection 2020. Biomed Res Int. 2020. PMID: 32908873 Free PMC article. - Estimating recombination rates from population-genetic data.
Stumpf MP, McVean GA. Stumpf MP, et al. Nat Rev Genet. 2003 Dec;4(12):959-68. doi: 10.1038/nrg1227. Nat Rev Genet. 2003. PMID: 14631356 Review. - Genetics in geographically structured populations: defining, estimating and interpreting F(ST).
Holsinger KE, Weir BS. Holsinger KE, et al. Nat Rev Genet. 2009 Sep;10(9):639-50. doi: 10.1038/nrg2611. Nat Rev Genet. 2009. PMID: 19687804 Free PMC article. Review.
Cited by
- Adaptive evolution: evaluating empirical support for theoretical predictions.
Olson-Manning CF, Wagner MR, Mitchell-Olds T. Olson-Manning CF, et al. Nat Rev Genet. 2012 Dec;13(12):867-77. doi: 10.1038/nrg3322. Nat Rev Genet. 2012. PMID: 23154809 Free PMC article. Review. - Genomics and the future of conservation genetics.
Allendorf FW, Hohenlohe PA, Luikart G. Allendorf FW, et al. Nat Rev Genet. 2010 Oct;11(10):697-709. doi: 10.1038/nrg2844. Nat Rev Genet. 2010. PMID: 20847747 Review. - Sex Differences in Recombination in Sticklebacks.
Sardell JM, Cheng C, Dagilis AJ, Ishikawa A, Kitano J, Peichel CL, Kirkpatrick M. Sardell JM, et al. G3 (Bethesda). 2018 May 31;8(6):1971-1983. doi: 10.1534/g3.118.200166. G3 (Bethesda). 2018. PMID: 29632132 Free PMC article. - Efficient detection and characterization of targets of natural selection using transfer learning.
Arnab SP, Dos Santos ALC, Fumagalli M, DeGiorgio M. Arnab SP, et al. bioRxiv [Preprint]. 2025 Mar 6:2025.03.05.641710. doi: 10.1101/2025.03.05.641710. bioRxiv. 2025. PMID: 40093065 Free PMC article. Updated. Preprint. - Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis.
Thomas CG, Wang W, Jovelin R, Ghosh R, Lomasko T, Trinh Q, Kruglyak L, Stein LD, Cutter AD. Thomas CG, et al. Genome Res. 2015 May;25(5):667-78. doi: 10.1101/gr.187237.114. Epub 2015 Mar 17. Genome Res. 2015. PMID: 25783854 Free PMC article.
References
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. - PubMed
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous