Initial genomics of the human nucleolus - PubMed (original) (raw)
Initial genomics of the human nucleolus
Attila Németh et al. PLoS Genet. 2010.
Abstract
We report for the first time the genomics of a nuclear compartment of the eukaryotic cell. 454 sequencing and microarray analysis revealed the pattern of nucleolus-associated chromatin domains (NADs) in the linear human genome and identified different gene families and certain satellite repeats as the major building blocks of NADs, which constitute about 4% of the genome. Bioinformatic evaluation showed that NAD-localized genes take part in specific biological processes, like the response to other organisms, odor perception, and tissue development. 3D FISH and immunofluorescence experiments illustrated the spatial distribution of NAD-specific chromatin within interphase nuclei and its alteration upon transcriptional changes. Altogether, our findings describe the nature of DNA sequences associated with the human nucleolus and provide insights into the function of the nucleolus in genome organization and establishment of nuclear architecture.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
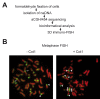
Figure 1. Genome-wide analyis of nucleolus-associated DNA.
(A) Experimental strategy. (B) 2D FISH analysis of nucleolus-associated DNA on human female lymphocyte metaphase spreads in the absence (-Cot1) or presence (+Cot1) of Cot1 competitor DNA. Arrows indicate chromosome 1 centromeres, arrowheads indicate p-arms of acrocentric chromosomes.
Figure 2. Genomic and size distribution of NADs.
(A) Distribution of NADs together with satellite repeats along human chromosomes. Note that the p-arms of the five acrocentric chromosomes (13, 14, 15, 21 and 22) were not analysed because they are not assembled in the hg18 genome build. NADs are labeled with red, satellite repeats with deep blue, centromeres with yellow and chromosomes with light blue (B) Histogram of NAD sizes; median = 749 kb; a total of 97 NADs were identified.
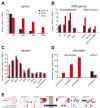
Figure 3. Sequence and chromatin features of NADs.
(A) RefSeq gene (B) RNA gene and (C) repeat statistics of NADs, genome and LADs. ZNF, OR and DEF indicate zinc finger, olfactory receptor and defensin gene families, respectively. RNA gene analysis of the ‘RepeatMasker’ and ‘RNA Genes’ tracks of the UCSC Genome Browser are shown on the left and right, respectively. (D) Chromatin features of NADs. Enrichment of functionally characterised repressive histone marks H3K27Me3, H3K9Me3 and H4K20Me3 in NADs are shown on the left, whereas depletion of the active histone mark H3K4Me1 is shown on the right. Genome, NADs and LADs values are labeled uniformly in (A–D) with black, red and white, respectively. The complete analysis is summarised in Table S5 and S6. (E) NADs and their typical genomic features on chromosome 19. Brown rectangle indicates the centromere. Abbreviations: UR (Universität Regensburg) NADs – nucleolus-associated chromatin domains identified in this study, PolI pseudo –pseudogenes of RNA polymerase I transcribed rRNA genes, OR – olfactory receptor genes, ZNF – zinc finger genes, tRNA – transfer RNA genes (and pseudogenes) transcribed by RNA polymerase III, NKI (Nederlands Kanker Instituut) LADs–lamin-associated chromosome domains identified in the Tig3 cell line .
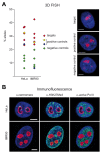
Figure 4. 3D immuno–FISH analysis of nucleolus-associated chromatin domains.
(A) Histograms show the frequency of the nucleolar localisation of NADs and control chromosomal regions detected by 3D FISH in HeLa cervix carcinoma and IMR90 diploid fibroblast cells. Percentage of nucleolus-associated alleles is shown on the left. Red diamond indicates target, green ones negative controls, whereas yellow diamond indicates the chromosome X pericentromeric and blue diamond the 5S cluster positive controls, respectively (see Table S7 for further BAC details). Single light optical sections of HeLa nuclei are shown on the right. BAC hybridization signals of RP11-90G23 target, RP5-915N17 positive control and RP11-81M8 negative control BACs are shown in green, nucleolar staining in red and DAPI counterstain in blue (scale bars: 5 µm). (B) α-H3K27Me3, α-centromere, α-active Pol II and α-B23/nucleophosmin immunostaining of HeLa and IMR90 cells. α-H3K27Me3, α-centromere and α-active Pol II signals are shown in green, nucleolar staining in red and DAPI counterstain in blue (scale bars: 5 µm).
Similar articles
- A Method to Identify Nucleolus-Associated Chromatin Domains (NADs).
Carpentier MC, Picart-Picolo A, Pontvianne F. Carpentier MC, et al. Methods Mol Biol. 2018;1675:99-109. doi: 10.1007/978-1-4939-7318-7_7. Methods Mol Biol. 2018. PMID: 29052188 - Methods for mapping 3D-chromosome architecture around nucleoli.
Bersaglieri C, Santoro R. Bersaglieri C, et al. Curr Opin Cell Biol. 2023 Apr;81:102171. doi: 10.1016/j.ceb.2023.102171. Epub 2023 May 23. Curr Opin Cell Biol. 2023. PMID: 37230037 Review. - Genome Organization in and around the Nucleolus.
Bersaglieri C, Santoro R. Bersaglieri C, et al. Cells. 2019 Jun 12;8(6):579. doi: 10.3390/cells8060579. Cells. 2019. PMID: 31212844 Free PMC article. Review. - Grabbing the genome by the NADs.
Matheson TD, Kaufman PD. Matheson TD, et al. Chromosoma. 2016 Jun;125(3):361-71. doi: 10.1007/s00412-015-0527-8. Epub 2015 Jul 15. Chromosoma. 2016. PMID: 26174338 Free PMC article. Review. - Genome-wide maps of nucleolus interactions reveal distinct layers of repressive chromatin domains.
Bersaglieri C, Kresoja-Rakic J, Gupta S, Bär D, Kuzyakiv R, Panatta M, Santoro R. Bersaglieri C, et al. Nat Commun. 2022 Mar 18;13(1):1483. doi: 10.1038/s41467-022-29146-2. Nat Commun. 2022. PMID: 35304483 Free PMC article.
Cited by
- Chromatin tracing and multiplexed imaging of nucleome architectures (MINA) and RNAs in single mammalian cells and tissue.
Liu M, Yang B, Hu M, Radda JSD, Chen Y, Jin S, Cheng Y, Wang S. Liu M, et al. Nat Protoc. 2021 May;16(5):2667-2697. doi: 10.1038/s41596-021-00518-0. Epub 2021 Apr 26. Nat Protoc. 2021. PMID: 33903756 Free PMC article. - Systematic characterization of the conformation and dynamics of budding yeast chromosome XII.
Albert B, Mathon J, Shukla A, Saad H, Normand C, Léger-Silvestre I, Villa D, Kamgoue A, Mozziconacci J, Wong H, Zimmer C, Bhargava P, Bancaud A, Gadal O. Albert B, et al. J Cell Biol. 2013 Jul 22;202(2):201-10. doi: 10.1083/jcb.201208186. J Cell Biol. 2013. PMID: 23878273 Free PMC article. - Mechanical regulation of nuclear structure and function.
Martins RP, Finan JD, Guilak F, Lee DA. Martins RP, et al. Annu Rev Biomed Eng. 2012;14:431-55. doi: 10.1146/annurev-bioeng-071910-124638. Epub 2012 May 22. Annu Rev Biomed Eng. 2012. PMID: 22655599 Free PMC article. Review. - The hierarchy of the 3D genome.
Gibcus JH, Dekker J. Gibcus JH, et al. Mol Cell. 2013 Mar 7;49(5):773-82. doi: 10.1016/j.molcel.2013.02.011. Mol Cell. 2013. PMID: 23473598 Free PMC article. Review. - Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability.
Yang L, Song T, Chen L, Kabra N, Zheng H, Koomen J, Seto E, Chen J. Yang L, et al. Mol Cell Biol. 2013 Oct;33(19):3835-48. doi: 10.1128/MCB.00476-13. Epub 2013 Jul 29. Mol Cell Biol. 2013. PMID: 23897426 Free PMC article.
References
- Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, et al. Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis. Cancer Res 2009 - PubMed
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
- Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources