Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers - PubMed (original) (raw)
Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers
Valer Gotea et al. Genome Res. 2010 May.
Abstract
Clustering of multiple transcription factor binding sites (TFBSs) for the same transcription factor (TF) is a common feature of cis-regulatory modules in invertebrate animals, but the occurrence of such homotypic clusters of TFBSs (HCTs) in the human genome has remained largely unknown. To explore whether HCTs are also common in human and other vertebrates, we used known binding motifs for vertebrate TFs and a hidden Markov model-based approach to detect HCTs in the human, mouse, chicken, and fugu genomes, and examined their association with cis-regulatory modules. We found that evolutionarily conserved HCTs occupy nearly 2% of the human genome, with experimental evidence for individual TFs supporting their binding to predicted HCTs. More than half of the promoters of human genes contain HCTs, with a distribution around the transcription start site in agreement with the experimental data from the ENCODE project. In addition, almost half of the 487 experimentally validated developmental enhancers contain them as well--a number more than 25-fold larger than expected by chance. We also found evidence of negative selection acting on TFBSs within HCTs, as the conservation of TFBSs is stronger than the conservation of sequences separating them. The important role of HCTs as components of developmental enhancers is additionally supported by a strong correlation between HCTs and the binding of the enhancer-associated coactivator protein Ep300 (also known as p300). Experimental validation of HCT-containing elements in both zebrafish and mouse suggest that HCTs could be used to predict both the presence of enhancers and their tissue specificity, and are thus a feature that can be effectively used in deciphering the gene regulatory code. In conclusion, our results indicate that HCTs are a pervasive feature of human cis-regulatory modules and suggest that they play an important role in gene regulation in the human and other vertebrate genomes.
Figures
Figure 1.
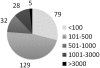
The distribution of the HCTs abundance for the 273 PWMs. The number of PWMs with abundant HCTs (>1000) is limited to 33, the best represented TFs being E2F1 (PWM: V$E2F_Q2; 5194 HCTs), ZFP161 (V$ZF5_01; 4844), and TEAD2 (V$ETF_Q6; 4382).
Figure 2.
Coverage of protein-coding genes by E2F HCTs. The coverage profile reveals a clear peak with a symmetric distribution around the transcription start site (TSS). Gene features are represented proportional to the median values of all human protein coding genes: 5′ UTR, 181 nt; first coding exon, 124; first intron, 2682; internal coding exon, 123 (multiple internal exons, as well as introns, are pooled together); internal intron, 1419; last coding exon, 149; 3′ UTR, 751. The promoter region is considered to be 1.5 kb in all cases. Depending on the length of the various gene features at a particular locus, HCTs could occasionally reach internal introns and exons. In the case of genes with split 5′ UTR regions, the first intron is likely to be covered by a promoter-based HCT, resulting, as in this case, in a higher coverage relative to the first coding exon.
Figure 3.
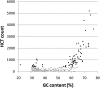
Bimodal distribution of the HCT count for 273 PWMs with respect to the GC content of HCTs. PWMs with more than 400 HCTs (28.6%, black) have HCTs that are either AT-rich (5.5%) or GC-rich (23.1%). The GC content was calculated for the entire span of the HCT, but avoiding coding and repetitive sequences.
Figure 4.
GC content, SNP density, and human–chimp divergence profiles of 5194 E2F1 HCTs. The GC content and the divergence from chimp increase, while the SNP density decreases sharply in HCTs as compared to flanking regions, which show values similar to the genome average. The increased GC content at the beginning and end of the HCT is due to the first and last E2F1 TFBSs that define the HCT, whereas the position of the other TFBSs is variable. All values were computed by avoiding coding and repetitive sequences.
Figure 5.
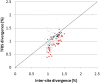
Divergence in TFBSs and intersite sequences within HCTs for 273 PWMs. The sequence divergence in human–chimp comparisons indicates that TFBSs tend to be more conserved than intersite sequences for HCTs of most PWMs (86.4%). For 31 of them (red) the difference is significant (after correction for multiple testing).
Figure 6.
Developmental enhancers overlap conserved HCTs. (A) The fraction of enhancers functional in mouse (487) that overlap with at least three sites in HCTs (39.2%, yellow) is significantly higher (P < 8.5 × 10−102) than what is expected by chance (1.5%, black). The fraction increases to above 50% if a lower number of TFBSs in an HCT are allowed for an HCT-enhancer overlap. (B) Coverage of positive enhancers by HCTs shows that HCTs are located toward the center of the enhancer. (C) HCTs of different transcription factors are associated with enhancers that show tissue specific, e.g., heart (A), hindbrain (B), or forebrain (D), or a more ubiquitous expression pattern (C,E) in E11.5 mouse embryos. The shade of gray is proportional to the number of tissue-specific enhancers overlapping HCTs for a specific TF, with the lightest shade of gray indicating one, and black indicating 10 (only observed for NOBOX and forebrain-specific enhancers) overlap instances. The tissues and the corresponding number of enhancers found to be active in them (indicated in parentheses) are: Mesenchyme derived from neural crest (1), facial mesenchyme (6), heart (20), other (22), cranial nerve (25), trigeminal V (ganglion, cranial) (18), dorsal root ganglion (34), limb (79), hindbrain (rhombencephalon) (142), midbrain (mesencephalon) (148), forebrain (156), neural tube (103), branchial arch (22), nose (20), eye (32), melanocytes (3), genital tubercle (3), somite (7), ear (6), and tail (2).
Figure 7.
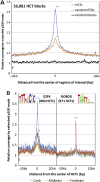
The enhancer activity of HCTs is supported by their association with the enhancer coactivator protein Ep300 in mouse. (A) Mouse HCTs corresponding to all human HCTs for 273 PWMs were combined into 56,081 regions (median size, 549 bp). Their Ep300 coverage profile was constructed by overlapping a total of 9,519,543 Ep300 reads with 40-kb regions centered on the middle point of HCTs. Even though ECRs are known to be significantly associated with Ep300 (P = 0), HCTs show an even stronger association by the means of a significantly higher Ep300 peak (P = 0). (B) Ep300 coverage profiles for HCTs of specific TFs and tissues reveal tissue-specific activity for certain TFs. For example, the coverage profile of 683 E2F4 HCTs reveals a peak significantly higher in limb than in either forebrain (P = 1.4 × 10−21) or midbrain (P = 1.2 × 10−40), while the difference between the forebrain and midbrain coverage is only marginally significant (P = 9.7 × 10−6). In the case of the 971 NOBOX HCTs, their coverage is significantly higher in forebrain than in either limb (P = 2.7 × 10−27) or midbrain (P = 7.3 × 10−43), while limb and midbrain coverage are not significantly different from each other (P = 0.07). These data strongly suggest a limb-specific activity for E2F4 HCTs, and a forebrain-specific activity for NOBOX HCTs. Statistical significance was evaluated with Fisher's exact test, as in Visel et al. (2009);*P < 0.01; **P < 10−20.
Figure 8.
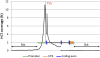
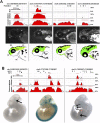
Experimental validation of predicted enhancers. (A) Four constructs containing POU3F2 HCTs that produced reproducible expression patterns of GFP in zebrafish. The sequence conservation profiles for mouse, chicken, and zebrafish represented as ECR Browser screen shots, correspond to the entire amplified regions (Table 6). Positions of the POU3F2 binding sites are represented by short vertical black bars above the conservation profile. All four elements are represented at the same scale. Pictures of 48–72-h post-fertilization zebrafish embryos with representative GFP expression patterns are shown below each element. The corresponding spatial domains of expression of each enhancer are also diagrammatically illustrated (regions where enhancer activity was recorded are shown in green). FB, forebrain; MB, midbrain; HB, hindbrain; E, eye; OC, otic capsule; HE, heart; YS, yolk sac. (B) Three constructs containing HCTs for NRF1 and E2F4 that produced reproducible LacZ expression pattern in transgenic mice. All three elements are represented at the same scale (this is different from the scale of elements tested in zebrafish). Arrows point to specific organs where the activity of these enhancers was observed, namely diencephalon for the enhancer on chromosome 1, and pancreas and caudal somites for the enhancer located in chromosome 10. Additional replicates for all elements presented here are included in the Supplemental material.
Similar articles
- Probing transcription factor combinatorics in different promoter classes and in enhancers.
Vandel J, Cassan O, Lèbre S, Lecellier CH, Bréhélin L. Vandel J, et al. BMC Genomics. 2019 Feb 1;20(1):103. doi: 10.1186/s12864-018-5408-0. BMC Genomics. 2019. PMID: 30709337 Free PMC article. - ConBind: motif-aware cross-species alignment for the identification of functional transcription factor binding sites.
Lelieveld SH, Schütte J, Dijkstra MJ, Bawono P, Kinston SJ, Göttgens B, Heringa J, Bonzanni N. Lelieveld SH, et al. Nucleic Acids Res. 2016 May 5;44(8):e72. doi: 10.1093/nar/gkv1518. Epub 2015 Dec 31. Nucleic Acids Res. 2016. PMID: 26721389 Free PMC article. - Genome-wide prediction of transcriptional regulatory elements of human promoters using gene expression and promoter analysis data.
Kim SY, Kim Y. Kim SY, et al. BMC Bioinformatics. 2006 Jul 4;7:330. doi: 10.1186/1471-2105-7-330. BMC Bioinformatics. 2006. PMID: 16817975 Free PMC article. - New insights into promoter-enhancer communication mechanisms revealed by dynamic single-molecule imaging.
Li J, Pertsinidis A. Li J, et al. Biochem Soc Trans. 2021 Jun 30;49(3):1299-1309. doi: 10.1042/BST20200963. Biochem Soc Trans. 2021. PMID: 34060610 Free PMC article. Review. - Towards genome-wide prediction and characterization of enhancers in plants.
Marand AP, Zhang T, Zhu B, Jiang J. Marand AP, et al. Biochim Biophys Acta Gene Regul Mech. 2017 Jan;1860(1):131-139. doi: 10.1016/j.bbagrm.2016.06.006. Epub 2016 Jun 16. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27321818 Review.
Cited by
- Ultrasensitive response motifs: basic amplifiers in molecular signalling networks.
Zhang Q, Bhattacharya S, Andersen ME. Zhang Q, et al. Open Biol. 2013 Apr 24;3(4):130031. doi: 10.1098/rsob.130031. Open Biol. 2013. PMID: 23615029 Free PMC article. Review. - Dynamic transcription factor networks in epithelial-mesenchymal transition in breast cancer models.
Siletz A, Schnabel M, Kniazeva E, Schumacher AJ, Shin S, Jeruss JS, Shea LD. Siletz A, et al. PLoS One. 2013 Apr 8;8(4):e57180. doi: 10.1371/journal.pone.0057180. Print 2013. PLoS One. 2013. PMID: 23593114 Free PMC article. - Differences in local genomic context of bound and unbound motifs.
Hansen L, Mariño-Ramírez L, Landsman D. Hansen L, et al. Gene. 2012 Sep 10;506(1):125-34. doi: 10.1016/j.gene.2012.06.005. Epub 2012 Jun 10. Gene. 2012. PMID: 22692006 Free PMC article. - Systematic interrogation of human promoters.
Weingarten-Gabbay S, Nir R, Lubliner S, Sharon E, Kalma Y, Weinberger A, Segal E. Weingarten-Gabbay S, et al. Genome Res. 2019 Feb;29(2):171-183. doi: 10.1101/gr.236075.118. Epub 2019 Jan 8. Genome Res. 2019. PMID: 30622120 Free PMC article. - fcScan: a versatile tool to cluster combinations of sites using genomic coordinates.
El-Kurdi A, Khalil GA, Khazen G, Khoueiry P. El-Kurdi A, et al. BMC Bioinformatics. 2020 May 19;21(1):194. doi: 10.1186/s12859-020-3536-4. BMC Bioinformatics. 2020. PMID: 32429868 Free PMC article.
References
- Adachi N, Lieber MR 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807–809 - PubMed
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310 - PubMed
- Araki E, Murakami T, Shirotani T, Kanai F, Shinohara Y, Shimada F, Mori M, Shichiri M, Ebina Y 1991. A cluster of four Sp1 binding sites required for efficient expression of the human insulin receptor gene. J Biol Chem 266: 3944–3948 - PubMed
- Arnone MI, Davidson EH 1997. The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG003988/HG/NHGRI NIH HHS/United States
- HL088393/HL/NHLBI NIH HHS/United States
- HL066681/HL/NHLBI NIH HHS/United States
- HG003988/HG/NHGRI NIH HHS/United States
- R01 HG004428/HG/NHGRI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- HG004428/HG/NHGRI NIH HHS/United States
- R01 HL088393/HL/NHLBI NIH HHS/United States
- U01 HL066681/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous