Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite - PubMed (original) (raw)
Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite
Rebecca D Schulte et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The coevolution between hosts and parasites is predicted to have complex evolutionary consequences for both antagonists, often within short time periods. To date, conclusive experimental support for the predictions is available mainly for microbial host systems, but for only a few multicellular host taxa. We here introduce a model system of experimental coevolution that consists of the multicellular nematode host Caenorhabditis elegans and the microbial parasite Bacillus thuringiensis. We demonstrate that 48 host generations of experimental coevolution under controlled laboratory conditions led to multiple changes in both parasite and host. These changes included increases in the traits of direct relevance to the interaction such as parasite virulence (i.e., host killing rate) and host resistance (i.e., the ability to survive pathogens). Importantly, our results provide evidence of reciprocal effects for several other central predictions of the coevolutionary dynamics, including (i) possible adaptation costs (i.e., reductions in traits related to the reproductive rate, measured in the absence of the antagonist), (ii) rapid genetic changes, and (iii) an overall increase in genetic diversity across time. Possible underlying mechanisms for the genetic effects were found to include increased rates of genetic exchange in the parasite and elevated mutation rates in the host. Taken together, our data provide comprehensive experimental evidence of the consequences of host-parasite coevolution, and thus emphasize the pace and complexity of reciprocal adaptations associated with these antagonistic interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Schematic overview of experimental setup. (A) Treatment conditions were identical except for the presence or absence of an antagonist. In the host control (blue), C. elegans (squiggles) adapted to the general experimental conditions (20 replicate populations). In the coevolution treatment (red), surviving C. elegans hosts (squiggles) and nematode-killing B. thuringiensis parasites (dots) coevolved (20 replicates). In the parasite control (green), pathogenic B. thuringiensis adapted to the general experimental conditions in the absence of nematodes (10 replicates). (B) After 48 host generations (i.e., end of experimental evolution), phenotypic and genetic effects were assayed. Phenotypes were compared among random pairs of coevolution and control replicates. For resistance/virulence analysis, each coevolution/control pair was exposed to an identical replicate population of the antagonist. Genetic analyses were conducted on DNA from 10 parasite clones per replicate and 20 host lines per replicate. (C) For genetic analysis over time, pooled population DNA samples were harvested from each replicate every fourth host generation. (D) Scanning electron micrograph of the front half of an infected nematode, broken open at the middle, shows internal disintegration and the presence of vegetative B. thuringiensis. Inset: higher magnification of the opened section.
Fig. 2.
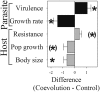
Phenotypic consequences of 48 host generations of experimental evolution. Phenotypes examined for parasite (black) and host (gray) included parasite virulence, parasite growth rate, host resistance, host population growth rate, and host body size. The bars represent the mean differences (±SEM) between randomly paired replicates from the coevolution and control treatments, calculated by subtracting the control values from the corresponding coevolution values (all values standardized to ensure comparability). Statistical analyses of the original paired data revealed that coevolution leads to significantly higher parasite virulence (paired t test, t = 3.53, P = 0.006), a trend toward higher host resistance (t = 1.93, P = 0.070), significantly lower parasite growth rate (t = −4.61, P < 0.001), significantly smaller host body size (t = −2.33, P = 0.032), and a trend toward lower host population growth rate (t = −1.93, P = 0.070). Significant results are indicated by asterisks and trends by asterisks in brackets.
Fig. 3.
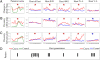
Evolution across time. Temporal changes in allele frequency/gene prevalence (A), gene diversity within replicate populations (B), and gene diversity between replicate populations within treatments (C), resulting in three main evolutionary patterns (D). We determined the relative prevalence of three toxin genes in each B. thuringiensis (BT) population and the relative allele frequencies in nine microsatellites in each C. elegans (CE) population. Change in gene/allele frequency was calculated as the absolute frequency difference between two subsequent sampling points averaged across genes/alleles for each replicate population. Gene diversity was calculated for each time point from relative gene/allele frequencies. At the beginning of experimental evolution (host generation 0), all populations of the various treatments had identical genotype mixtures and thus diversity levels. Points represent means per sampling point (±SEM) for the combined parasite toxin genes, for all nine host loci combined, and for four representative host loci separately (II-R, 4001, IV-L, and V-L). Red asterisks indicate significantly larger values for coevolution conditions [general linear model (GLM), likelihood ratio test for treatment, χ2 ≥ 5.71, P ≤ 0.017; detailed results in
Table S1
); blue asterisks for control conditions (χ2 ≥ 5.55, P ≤ 0.018). The remaining two graphs show insignificant differences (χ2 ≤ 0.12, P ≥ 0.730). The analyses identified three patterns whereby host–parasite coevolution was associated with an increase in evolutionary rates and diversities (pattern I), an increase in evolutionary rates and diversities across populations but decreased within-population diversities (pattern II), or a decrease in diversities and no difference in evolutionary rates (pattern III).
Fig. 4.
Snap-shot look at possible diversity-generating mechanisms after 48 host generations of experimental evolution. (A) Genetic exchange in B. thuringiensis (average number of clones with more than one toxin gene per replicate) was significantly higher upon coevolution (Wilcoxon test, Z = −3.01, P = 0.003). Bars represent means (±SEM). (B) Host mutations in C. elegans (average number of mutated loci per replicate) tended to be more frequent after coevolution (Wilcoxon test, Z = 2.45, P = 0.02, significance level adjusted to α = 0.01 according to FDR;
Table S1
).
Similar articles
- Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer.
Schulte RD, Makus C, Schulenburg H. Schulte RD, et al. J Evol Biol. 2013 Aug;26(8):1836-40. doi: 10.1111/jeb.12174. Epub 2013 Jul 19. J Evol Biol. 2013. PMID: 23865952 - Host-parasite local adaptation after experimental coevolution of Caenorhabditis elegans and its microparasite Bacillus thuringiensis.
Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Schulte RD, et al. Proc Biol Sci. 2011 Sep 22;278(1719):2832-9. doi: 10.1098/rspb.2011.0019. Epub 2011 Feb 9. Proc Biol Sci. 2011. PMID: 21307053 Free PMC article. - The role of defensive symbionts in host-parasite coevolution.
Vorburger C, Perlman SJ. Vorburger C, et al. Biol Rev Camb Philos Soc. 2018 Nov;93(4):1747-1764. doi: 10.1111/brv.12417. Epub 2018 Apr 16. Biol Rev Camb Philos Soc. 2018. PMID: 29663622 Review. - Novel genomic approaches to study antagonistic coevolution between hosts and parasites.
Märkle H, John S, Cornille A, Fields PD, Tellier A. Märkle H, et al. Mol Ecol. 2021 Aug;30(15):3660-3676. doi: 10.1111/mec.16001. Epub 2021 Jun 22. Mol Ecol. 2021. PMID: 34038012 Review.
Cited by
- The evolution of coexistence from competition in experimental co-cultures of Escherichia coli and Saccharomyces cerevisiae.
Barber JN, Sezmis AL, Woods LC, Anderson TD, Voss JM, McDonald MJ. Barber JN, et al. ISME J. 2021 Mar;15(3):746-761. doi: 10.1038/s41396-020-00810-z. Epub 2020 Oct 22. ISME J. 2021. PMID: 33093620 Free PMC article. - Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis.
Boehnisch C, Wong D, Habig M, Isermann K, Michiels NK, Roeder T, May RC, Schulenburg H. Boehnisch C, et al. PLoS One. 2011;6(9):e24619. doi: 10.1371/journal.pone.0024619. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931778 Free PMC article. - Cross-Resistance: A Consequence of Bi-partite Host-Parasite Coevolution.
Biswas T, Joop G, Rafaluk-Mohr C. Biswas T, et al. Insects. 2018 Feb 26;9(1):28. doi: 10.3390/insects9010028. Insects. 2018. PMID: 29495405 Free PMC article. - Testing GxG interactions between coinfecting microbial parasite genotypes within hosts.
Bose J, Schulte RD. Bose J, et al. Front Genet. 2014 May 14;5:124. doi: 10.3389/fgene.2014.00124. eCollection 2014. Front Genet. 2014. PMID: 24860594 Free PMC article. - Ecological effects of aphid abundance, genotypic variation, and contemporary evolution on plants.
Turley NE, Johnson MT. Turley NE, et al. Oecologia. 2015 Jul;178(3):747-59. doi: 10.1007/s00442-015-3276-8. Epub 2015 Mar 5. Oecologia. 2015. PMID: 25740334
References
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. - PubMed
- Thompson JN. The Geographic Mosaic of Coevolution. Chicago: Univ. of Chicago Press; 2005.
- Decaestecker E, et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. - PubMed
- Jokela J, Dybdahl MF, Lively CM. The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am Nat. 2009;174(Suppl 1):S43–S53. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources