A benchmark of parametric methods for horizontal transfers detection - PubMed (original) (raw)
A benchmark of parametric methods for horizontal transfers detection
Jennifer Becq et al. PLoS One. 2010.
Abstract
Horizontal gene transfer (HGT) has appeared to be of importance for prokaryotic species evolution. As a consequence numerous parametric methods, using only the information embedded in the genomes, have been designed to detect HGTs. Numerous reports of incongruencies in results of the different methods applied to the same genomes were published. The use of artificial genomes in which all HGT parameters are controlled allows testing different methods in the same conditions. The results of this benchmark concerning 16 representative parametric methods showed a great variety of efficiencies. Some methods work very poorly whatever the type of HGTs and some depend on the conditions or on the metrics used. The best methods in terms of total errors were those using tetranucleotides as criterion for the window methods or those using codon usage for gene based methods and the Kullback-Leibler divergence metric. Window methods are very sensitive but less specific and detect badly lone isolated gene. On the other hand gene based methods are often very specific but lack of sensitivity. We propose using two methods in combination to get the best of each category, a gene based one for specificity and a window based one for sensitivity.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
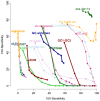
Figure 1. ROC-like curves of the 16 methods.
Each dot of a curve corresponds to the values of type I error (100-sensitivity) and type II error (100-specificity) for each value of r (see M&M). The best methods are those with the less errors, i.e. those that are the closest of the origin.
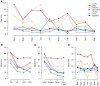
Figure 2. Mean errors of 7 methods according to (A) origin, (B) overall quantity, (C) size and (D) recipient genome.
The mean error is the mean of type I (sensitivity) and type II (specificity) errors. It is presented here for the 7 efficient HT detection methods of each criterion (codon usage: CU.KL; dinucleotide frequencies: dint5; GC content: GCtotal and GC1-GC3; and tetranucleotide frequencies: oli.chi2, oli.KL and signature) according to four parameters. A: the origin. The unique donor genome of the HTs are ordered according to their distance to the host genome (E. coli) in terms of tetranucleotide frequencies – the closest on the left and the farthest on the right. B: the overall quantity of HTs in percentage of the genome. C: the size of the HTs. Small, Medium, Large and Very Large respectively mean 1 to 5 genes, 5 to 10 genes, 10 to 20 genes and 20 to 30 genes. D: the host genome, i.e. the genome receiving the HTs.
References
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. - PubMed
- Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–8. - PubMed
- Dutta C, Pan A. Horizontal gene transfer and bacterial diversity. J Biosci. 2002;27:27–33. - PubMed
- Eisen JA. Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Current Opinion in Genetics & Development. 2000;10:606–611. - PubMed
- Gogarten JP, Doolittle WF, Lawrence J, G. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials