Histo-proteomic profiling of formalin-fixed, paraffin-embedded tissue - PubMed (original) (raw)
Review
Histo-proteomic profiling of formalin-fixed, paraffin-embedded tissue
Kant M Matsuda et al. Expert Rev Proteomics. 2010 Apr.
Abstract
In the functional proteome era, the proteomic profiling of clinicopathologic-annotated tissues is an essential step for mining and evaluating candidate biomarkers for disease. For many diseases, but especially cancer, the development of predictive biomarkers requires performing assays directly on the diseased tissue. The last decade has seen the explosion of both prognostic and predictive biomarkers in the research setting but few of these biomarkers have entered widespread clinical use. Previously, application of routine proteomic methodologies to clinical formalin-fixed and paraffin-embedded tissue specimens has provided unsatisfactory results. In this paper, we will discuss recent advancements in proteomic profiling technology for clinical applications. These approaches focus on the retention of histomorphologic information as an element of the proteomic analysis.
Conflict of interest statement
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Figures
Figure 1.. Components of multiplex tissue immunoblotting.
Diagram showing the transfer ‘package’ utilized for transfer of proteins from a glass slide to membranes of tissue immunoblotting. The package is assembled, then sealed in a Kapak SealPAK® pouch. The package is placed on a heat block and then applied to a multiple-serial heating system (1 h at 55°C, 0.5 h at 65°C and 2 h at 80°C). A weight applied to the top of the transfer assembly unit helps to ensure a tight connection between the layers of material used in the transfer system. The proteins are transferred from the tissue section and are deposited onto the membrane stack. After transfer, the package is disassembled, the membrane stack separated and the proteins on each membrane are individually probed by means of conventional protein blotting.
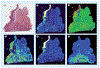
Figure 2.. Protein expression in progression from normal to invasive cancer as determined by tissue immunoblotting.
(A) Routine hematoxylin and eosin stain section of esophageal cancer (indicated as an area labeled with ‘T’) along with associated dysplastic (indicated as a triangle area labeled with ‘D’) and normal epithelium (indicated as a rectangle area labeled with ‘N’). (B) CK-4, (C) CK-14, (D) cyclooxygenase-1, (E) annexin 1 and (F) secreted protein acidic and rich in cysteine. Fluorescent scans represented with pseudocolor, where signal intensity is white–red–yellow–green–blue–black from maximum to minimum signal. Modified from [11].
Figure 3.. Correlation among high-resolution MRI-hisotopathology-multiplex tissue immunoblotting in identical planes.
(A) Ex vivo T2*-MRI of removed human breast tissue at in-plane resolution of 25 × 25 μm. (B) Routine hematoxylin and eosin section. (C) Multiplex tissue immunoblotting for HER2. High-intensity signal is expressed in red color.
Figure 4.. Schematic of potential application of multiplex tissue immunoblotting in correlation studies with molecular imaging.
Breast specimen is used as an example in this schematic presentation of the work-flow for the correlation study. The arrow indicates co-registration or overlay among radiological imaging (MRI, PET and CT), histopathology section (whole-mount section or regular-size section) and protein expression by MTIB. Initially, in vivo imaging, such as molecular PET imaging for the targeted molecule, is performed, and co-registered with other imaging studies (CT or MRI). Following surgical intervention, ex vivo imaging of the removed tissue is performed and co-registered with in vivo imaging. The targeted lesion, which is seen in in vivo imaging, will be obtained through image-guided sectioning/sampling for histopathologic examination. Once identical planes are obtained, protein expression can be demonstrated using MTIB and readily be correlated with in vivo molecular imaging. MTIB: Multiplex tissue immunoblotting.
Similar articles
- Transfer and multiplex immunoblotting of a paraffin embedded tissue.
Chung JY, Hewitt SM. Chung JY, et al. Methods Mol Biol. 2009;536:139-48. doi: 10.1007/978-1-59745-542-8_16. Methods Mol Biol. 2009. PMID: 19378053 - Proteomic expressional profiling of a paraffin-embedded tissue by multiplex tissue immunoblotting.
Chung JY, Hewitt SM. Chung JY, et al. Methods Mol Biol. 2015;1312:175-84. doi: 10.1007/978-1-4939-2694-7_21. Methods Mol Biol. 2015. PMID: 26044002 Free PMC article. - Laser Microdissection-Based Microproteomics of Formalin-Fixed and Paraffin-Embedded (FFPE) Tissues.
Longuespée R, Baiwir D, Mazzucchelli G, Smargiasso N, De Pauw E. Longuespée R, et al. Methods Mol Biol. 2018;1723:19-31. doi: 10.1007/978-1-4939-7558-7_2. Methods Mol Biol. 2018. PMID: 29344853 - Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues.
Steiner C, Ducret A, Tille JC, Thomas M, McKee TA, Rubbia-Brandt L, Scherl A, Lescuyer P, Cutler P. Steiner C, et al. Proteomics. 2014 Mar;14(4-5):441-51. doi: 10.1002/pmic.201300311. Proteomics. 2014. PMID: 24339433 Free PMC article. Review. - Proteomic studies of formalin-fixed paraffin-embedded tissues.
Giusti L, Lucacchini A. Giusti L, et al. Expert Rev Proteomics. 2013 Apr;10(2):165-77. doi: 10.1586/epr.13.3. Expert Rev Proteomics. 2013. PMID: 23573783 Review.
Cited by
- Proteomics and diabetic nephropathy: what have we learned from a decade of clinical proteomics studies?
Papale M, Di Paolo S, Vocino G, Rocchetti MT, Gesualdo L. Papale M, et al. J Nephrol. 2014 Jun;27(3):221-8. doi: 10.1007/s40620-014-0044-5. Epub 2014 Feb 25. J Nephrol. 2014. PMID: 24567069 Review. - Ex vivo tissue imaging for radiology-pathology correlation: a pilot study with a small bore 7-T MRI in a rare pigmented ganglioglioma exhibiting complex MR signal characteristics associated with melanin and hemosiderin.
Matsuda KM, Lopes-Calcas A, Honke ML, O'Brien-Moran Z, Buist R, West M, Martin M. Matsuda KM, et al. J Med Imaging (Bellingham). 2017 Jul;4(3):036001. doi: 10.1117/1.JMI.4.3.036001. Epub 2017 Sep 13. J Med Imaging (Bellingham). 2017. PMID: 28924575 Free PMC article. - ASAP─Automated Sonication-Free Acid-Assisted Proteomes─from Cells and FFPE Tissues.
Barnabas GD, Goebeler V, Tsui J, Bush JW, Lange PF. Barnabas GD, et al. Anal Chem. 2023 Feb 14;95(6):3291-3299. doi: 10.1021/acs.analchem.2c04264. Epub 2023 Feb 1. Anal Chem. 2023. PMID: 36724070 Free PMC article. - Comparison of fixation methods for preservation of morphology, RNAs, and proteins from paraffin-embedded human cancer cell-implanted mouse models.
Matsuda Y, Fujii T, Suzuki T, Yamahatsu K, Kawahara K, Teduka K, Kawamoto Y, Yamamoto T, Ishiwata T, Naito Z. Matsuda Y, et al. J Histochem Cytochem. 2011 Jan;59(1):68-75. doi: 10.1369/jhc.2010.957217. J Histochem Cytochem. 2011. PMID: 20940453 Free PMC article. - A Buffered Alcohol-Based Fixative for Histomorphologic and Molecular Applications.
Perry C, Chung JY, Ylaya K, Choi CH, Simpson A, Matsumoto KT, Smith WA, Hewitt SM. Perry C, et al. J Histochem Cytochem. 2016 Jul;64(7):425-40. doi: 10.1369/0022155416649579. Epub 2016 May 24. J Histochem Cytochem. 2016. PMID: 27221702 Free PMC article.
References
- Schena M, Shalon D, Davis RW et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270(5235), 368–371 (1995). - PubMed
- Parekh RB, Rohlff C. Post-translational modification of proteins and the discovery of new medicine. Curr. Opin. Biotechnol 8(6), 718–723 (1997). - PubMed
- Marcotte EM. Measuring the dynamics of the proteome. Genome Res. 11(2), 191–193 (2001). - PubMed
- Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol. 410, 400–415 (2006).
•• First description of tissue microarray for searching new biomarkers.
- Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol. 410, 400–415 (2006).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources