Recent developments in classical density modification - PubMed (original) (raw)
Recent developments in classical density modification
Kevin Cowtan. Acta Crystallogr D Biol Crystallogr. 2010 Apr.
Abstract
Classical density-modification techniques (as opposed to statistical approaches) offer a computationally cheap method for improving phase estimates in order to provide a good electron-density map for model building. The rise of statistical methods has lead to a shift in focus away from the classical approaches; as a result, some recent developments have not made their way into classical density-modification software. This paper describes the application of some recent techniques, including most importantly the use of prior phase information in the likelihood estimation of phase errors within a classical density-modification framework. The resulting software gives significantly better results than comparable classical methods, while remaining nearly two orders of magnitude faster than statistical methods.
Figures
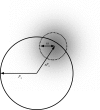
Figure 1
The terms F o, F c, s and ω describe the observed and calculated structure factor, the scale factor and the radius of the Gaussian error term in the Argand diagram. The shading represents the Gaussian probability distribution centred on sF c.
Figure 2
Mean map correlation calculated of a range of JCSG data sets with using different density-modification programs and options. (a) Parrot with no new features compared with DM. (b) Parrot with MLHL likelihood function compared with the Rice function. (c) As (b), with anisotropy correction compared with no anisotropy correction. (d) As (c), with automated NCS averaging compared with no NCS averaging.
Similar articles
- Automated structure solution, density modification and model building.
Terwilliger TC. Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1937-40. doi: 10.1107/s0907444902016438. Epub 2002 Oct 21. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12393925 - Statistical density modification using local pattern matching.
Terwilliger TC. Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2003 Oct;59(Pt 10):1688-701. doi: 10.1107/s0907444903015142. Epub 2003 Sep 19. Acta Crystallogr D Biol Crystallogr. 2003. PMID: 14501107 Free PMC article. - Reciprocal-space solvent flattening.
Terwilliger TC. Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1863-71. doi: 10.1107/s0907444999010033. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10531484 Free PMC article. - Error estimation and bias correction in phase-improvement calculations.
Cowtan K. Cowtan K. Acta Crystallogr D Biol Crystallogr. 1999 Sep;55(Pt 9):1555-67. doi: 10.1107/s0907444999007416. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10489450 Review. - EDM-DEDM and protein crystal structure solution.
Caliandro R, Carrozzini B, Cascarano GL, Giacovazzo C, Mazzone AM, Siliqi D. Caliandro R, et al. Acta Crystallogr D Biol Crystallogr. 2009 May;65(Pt 5):477-84. doi: 10.1107/S0907444909008609. Epub 2009 Apr 18. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19390153 Review.
Cited by
- Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Böth D, Steiner EM, Stadler D, Lindqvist Y, Schnell R, Schneider G. Böth D, et al. Acta Crystallogr D Biol Crystallogr. 2013 Mar;69(Pt 3):432-41. doi: 10.1107/S0907444912049268. Epub 2013 Feb 16. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23519418 Free PMC article. - Crystallographic snapshot of cellulose synthesis and membrane translocation.
Morgan JL, Strumillo J, Zimmer J. Morgan JL, et al. Nature. 2013 Jan 10;493(7431):181-6. doi: 10.1038/nature11744. Epub 2012 Dec 9. Nature. 2013. PMID: 23222542 Free PMC article. - Structure of the catalytic domain of the colistin resistance enzyme MCR-1.
Stojanoski V, Sankaran B, Prasad BV, Poirel L, Nordmann P, Palzkill T. Stojanoski V, et al. BMC Biol. 2016 Sep 21;14(1):81. doi: 10.1186/s12915-016-0303-0. BMC Biol. 2016. PMID: 27655155 Free PMC article. - The first transmembrane region of complement component-9 acts as a brake on its self-assembly.
Spicer BA, Law RHP, Caradoc-Davies TT, Ekkel SM, Bayly-Jones C, Pang SS, Conroy PJ, Ramm G, Radjainia M, Venugopal H, Whisstock JC, Dunstone MA. Spicer BA, et al. Nat Commun. 2018 Aug 15;9(1):3266. doi: 10.1038/s41467-018-05717-0. Nat Commun. 2018. PMID: 30111885 Free PMC article. - Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Cuervo A, Fàbrega-Ferrer M, Machón C, Conesa JJ, Fernández FJ, Pérez-Luque R, Pérez-Ruiz M, Pous J, Vega MC, Carrascosa JL, Coll M. Cuervo A, et al. Nat Commun. 2019 Aug 20;10(1):3746. doi: 10.1038/s41467-019-11705-9. Nat Commun. 2019. PMID: 31431626 Free PMC article.
References
- Abrahams, J. P. (1997). Acta Cryst. D53, 371–376. - PubMed
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. - PubMed
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
- Caliandro, R., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C. & Siliqi, D. (2005). Acta Cryst. D61, 556–565. - PubMed
- Cowtan, K. (1999). Acta Cryst. D55, 1555–1567. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources