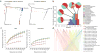QIIME allows analysis of high-throughput community sequencing data - PubMed (original) (raw)
doi: 10.1038/nmeth.f.303. Epub 2010 Apr 11.
Justin Kuczynski, Jesse Stombaugh, Kyle Bittinger, Frederic D Bushman, Elizabeth K Costello, Noah Fierer, Antonio Gonzalez Peña, Julia K Goodrich, Jeffrey I Gordon, Gavin A Huttley, Scott T Kelley, Dan Knights, Jeremy E Koenig, Ruth E Ley, Catherine A Lozupone, Daniel McDonald, Brian D Muegge, Meg Pirrung, Jens Reeder, Joel R Sevinsky, Peter J Turnbaugh, William A Walters, Jeremy Widmann, Tanya Yatsunenko, Jesse Zaneveld, Rob Knight
- PMID: 20383131
- PMCID: PMC3156573
- DOI: 10.1038/nmeth.f.303
QIIME allows analysis of high-throughput community sequencing data
J Gregory Caporaso et al. Nat Methods. 2010 May.
No abstract available
Conflict of interest statement
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/.
Figures
Figure 1
QIIME analyses of the distal gut microbiotas of conventionally raised and conventionalized mice, gnotobiotic mice colonized with a human fecal gut microbiota (H-mice), and human adult mono- and dizygotic twins. (a) Principal coordinates analysis plots for mice, H-mice and twins. Colors correspond to separate samples by species and time point, and are consistent throughout the panels. (b) Unweighted UniFrac distance histograms between the data for fecal microbiota of human twins; human donors for the H-mice study; day 56 post-transplant H-mice on a low-fat (LF) and plant polysaccharide–rich (PP) diet; day 1 H-mice (LF and PP diet); and day 0 H-mice. Taxonomic classifications are presented at the class level. (c) Alpha diversity rarefaction plots of phylogenetic diversity for the H-mice samples. (d) OTU network connectivity of H-mice time series data. CONV-D, conventionalized mice; CONV-R, conventionally raised mice; and GF, germ-free mice.
Similar articles
- Oral microbiota and dental caries data from monozygotic and dizygotic twin children.
Kasimoglu Y, Koruyucu M, Birant S, Karacan I, Topcuoglu N, Tuna EB, Gencay K, Seymen F. Kasimoglu Y, et al. Sci Data. 2020 Oct 13;7(1):348. doi: 10.1038/s41597-020-00691-z. Sci Data. 2020. PMID: 33051450 Free PMC article. - A method for differentiating between monozygotic and dizygotic twins.
Takahashi H, Goto S, Kobayashi M, Takeuchi S. Takahashi H, et al. Nihon Sanka Fujinka Gakkai Zasshi. 1989 Dec;41(12):2005-9. Nihon Sanka Fujinka Gakkai Zasshi. 1989. PMID: 2592823 - Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing.
Prodan A, Tremaroli V, Brolin H, Zwinderman AH, Nieuwdorp M, Levin E. Prodan A, et al. PLoS One. 2020 Jan 16;15(1):e0227434. doi: 10.1371/journal.pone.0227434. eCollection 2020. PLoS One. 2020. PMID: 31945086 Free PMC article. - Is optimism heritable? A study of twins.
Schulman P, Keith D, Seligman ME. Schulman P, et al. Behav Res Ther. 1993 Jul;31(6):569-74. doi: 10.1016/0005-7967(93)90108-7. Behav Res Ther. 1993. PMID: 8347115 Review. - [Laterality in twins].
Reiss M. Reiss M. Z Morphol Anthropol. 1996 Sep;81(2):141-55. Z Morphol Anthropol. 1996. PMID: 9312956 Review. German.
Cited by
- Microbial diversity of co-occurring heterotrophs in cultures of marine picocyanobacteria.
Kearney SM, Thomas E, Coe A, Chisholm SW. Kearney SM, et al. Environ Microbiome. 2021 Jan 6;16(1):1. doi: 10.1186/s40793-020-00370-x. Environ Microbiome. 2021. PMID: 33902739 Free PMC article. - A cross-taxon analysis of insect-associated bacterial diversity.
Jones RT, Sanchez LG, Fierer N. Jones RT, et al. PLoS One. 2013 Apr 16;8(4):e61218. doi: 10.1371/journal.pone.0061218. Print 2013. PLoS One. 2013. PMID: 23613815 Free PMC article. - Structure-constrained sparse canonical correlation analysis with an application to microbiome data analysis.
Chen J, Bushman FD, Lewis JD, Wu GD, Li H. Chen J, et al. Biostatistics. 2013 Apr;14(2):244-58. doi: 10.1093/biostatistics/kxs038. Epub 2012 Oct 15. Biostatistics. 2013. PMID: 23074263 Free PMC article. - Different Urban Forest Tree Species Affect the Assembly of the Soil Bacterial and Fungal Community.
Ao L, Zhao M, Li X, Sun G. Ao L, et al. Microb Ecol. 2022 Feb;83(2):447-458. doi: 10.1007/s00248-021-01754-3. Epub 2021 May 24. Microb Ecol. 2022. PMID: 34031701 - Diverse Diets with Consistent Core Microbiome in Wild Bee Pollen Provisions.
Dew RM, McFrederick QS, Rehan SM. Dew RM, et al. Insects. 2020 Aug 4;11(8):499. doi: 10.3390/insects11080499. Insects. 2020. PMID: 32759653 Free PMC article.
References
- Hopkin M. Nature. 2006;444:420–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG4872/HG/NHGRI NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- U01 HG004866-01/HG/NHGRI NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- GM65103/GM/NIGMS NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- GM8759/GM/NIGMS NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- HG4866/HG/NHGRI NIH HHS/United States
- LM9451/LM/NLM NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- DK83981/DK/NIDDK NIH HHS/United States
- UH2 DK083981/DK/NIDDK NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- UH3 DK083981/DK/NIDDK NIH HHS/United States
- R01 HG004872-01/HG/NHGRI NIH HHS/United States
- T15 LM009451/LM/NLM NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources