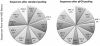Increased throughput by parallelization of library preparation for massive sequencing - PubMed (original) (raw)
Increased throughput by parallelization of library preparation for massive sequencing
Sverker Lundin et al. PLoS One. 2010.
Abstract
Background: Massively parallel sequencing systems continue to improve on data output, while leaving labor-intensive library preparations a potential bottleneck. Efforts are currently under way to relieve the crucial and time-consuming work to prepare DNA for high-throughput sequencing.
Methodology/principal findings: In this study, we demonstrate an automated parallel library preparation protocol using generic carboxylic acid-coated superparamagnetic beads and polyethylene glycol precipitation as a reproducible and flexible method for DNA fragment length separation. With this approach the library preparation for DNA sequencing can easily be adjusted to a desired fragment length. The automated protocol, here demonstrated using the GS FLX Titanium instrument, was compared to the standard manual library preparation, showing higher yield, throughput and great reproducibility. In addition, 12 libraries were prepared and uniquely tagged in parallel, and the distribution of sequence reads between these indexed samples could be improved using quantitative PCR-assisted pooling.
Conclusions/significance: We present a novel automated procedure that makes it possible to prepare 36 indexed libraries per person and day, which can be increased to up to 96 libraries processed simultaneously. The yield, speed and robust performance of the protocol constitute a substantial improvement to present manual methods, without the need of extensive equipment investments. The described procedure enables a considerable efficiency increase for small to midsize sequencing centers.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
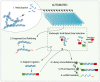
Figure 1. A schematic view of the automated process.
Step 1–6 is the regular reaction steps of library preparation. Each sample purification is shown with roman numerals and illustrated as arrows crossing the carboxylic acid beads size selection box.
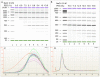
Figure 2. Length dependent precipitation of DNA.
DNA fragment length precipitation was controlled by varying final PEG concentration (as shown at the top), while NaCl concentration was kept constant at 0.9 M. As the PEG concentration rises, smaller fragments are precipitated. A: A DNA marker ranging from 100–10,000 bp are precipitated. B: A DNA marker ranging from 25–700 bp are precipitated. C: Nebulized DNA is size-selected using CA-beads and 8.3% PEG. Samples are analyzed using Bioanalyzer DNA 7500 kit and viewed using the Bioanalyzer software, where red curve show non-size selected nebulized sample and the other colors show 11 size-selected samples. D: Three libraries are prepared in parallel from nebulized C.tentans DNA, analyzed using Bioanalyzer RNA 6000 Pico kit and viewed using the Bioanalyzer software illustrating reproducibility.
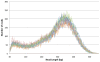
Figure 3. Read length distribution of the different library preparations.
Fragment length is plotted against occurrence of that length. All preparations show a similar pattern.
Figure 4. Equalizing indexed libraries.
Read distribution between MID-libraries generated from the improved automatic protocol, pooled according to Bioanalyzer (left) and qPCR concentration measurements (right). The developed protocol successfully produced evenly distributed barcoded reads, except for MID3 that consistently underperformed.
Similar articles
- Scalable transcriptome preparation for massive parallel sequencing.
Stranneheim H, Werne B, Sherwood E, Lundeberg J. Stranneheim H, et al. PLoS One. 2011;6(7):e21910. doi: 10.1371/journal.pone.0021910. Epub 2011 Jul 7. PLoS One. 2011. PMID: 21760920 Free PMC article. - Large scale library generation for high throughput sequencing.
Borgström E, Lundin S, Lundeberg J. Borgström E, et al. PLoS One. 2011 Apr 27;6(4):e19119. doi: 10.1371/journal.pone.0019119. PLoS One. 2011. PMID: 21589638 Free PMC article. - Methods for generating shotgun and mixed shotgun/paired-end libraries for the 454 DNA sequencer.
Wiley G, Macmil S, Qu C, Wang P, Xing Y, White D, Li J, White JD, Domingo A, Roe BA. Wiley G, et al. Curr Protoc Hum Genet. 2009 Apr;Chapter 18:Unit18.1. doi: 10.1002/0471142905.hg1801s61. Curr Protoc Hum Genet. 2009. PMID: 19360698 - Library preparation for next generation sequencing: A review of automation strategies.
Hess JF, Kohl TA, Kotrová M, Rönsch K, Paprotka T, Mohr V, Hutzenlaub T, Brüggemann M, Zengerle R, Niemann S, Paust N. Hess JF, et al. Biotechnol Adv. 2020 Jul-Aug;41:107537. doi: 10.1016/j.biotechadv.2020.107537. Epub 2020 Mar 19. Biotechnol Adv. 2020. PMID: 32199980 Review. - Library preparation methods for next-generation sequencing: tone down the bias.
van Dijk EL, Jaszczyszyn Y, Thermes C. van Dijk EL, et al. Exp Cell Res. 2014 Mar 10;322(1):12-20. doi: 10.1016/j.yexcr.2014.01.008. Epub 2014 Jan 15. Exp Cell Res. 2014. PMID: 24440557 Review.
Cited by
- Genome sequence of segmented filamentous bacteria present in the human intestine.
Jonsson H, Hugerth LW, Sundh J, Lundin E, Andersson AF. Jonsson H, et al. Commun Biol. 2020 Sep 4;3(1):485. doi: 10.1038/s42003-020-01214-7. Commun Biol. 2020. PMID: 32887924 Free PMC article. - Global gene expression analysis in etiolated and de-etiolated seedlings in conifers.
Ranade SS, Delhomme N, García-Gil MR. Ranade SS, et al. PLoS One. 2019 Jul 5;14(7):e0219272. doi: 10.1371/journal.pone.0219272. eCollection 2019. PLoS One. 2019. PMID: 31276530 Free PMC article. - Periodontal Health and Oral Microbiota in Patients with Rheumatoid Arthritis.
Eriksson K, Fei G, Lundmark A, Benchimol D, Lee L, Hu YOO, Kats A, Saevarsdottir S, Catrina AI, Klinge B, Andersson AF, Klareskog L, Lundberg K, Jansson L, Yucel-Lindberg T. Eriksson K, et al. J Clin Med. 2019 May 8;8(5):630. doi: 10.3390/jcm8050630. J Clin Med. 2019. PMID: 31072030 Free PMC article. - Rapid flow-sorting to simultaneously resolve multiplex massively parallel sequencing products.
Sandberg J, Werne B, Dessing M, Lundeberg J. Sandberg J, et al. Sci Rep. 2011;1:108. doi: 10.1038/srep00108. Epub 2011 Oct 6. Sci Rep. 2011. PMID: 22355625 Free PMC article. - Low-cost and High-throughput RNA-seq Library Preparation for Illumina Sequencing from Plant Tissue.
Bjornson M, Kajala K, Zipfel C, Ding P. Bjornson M, et al. Bio Protoc. 2020 Oct 20;10(20):e3799. doi: 10.21769/BioProtoc.3799. eCollection 2020 Oct 20. Bio Protoc. 2020. PMID: 33659453 Free PMC article.
References
- Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93:105–111. - PubMed
- Nyren P, Pettersson B, Uhlen M. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal Biochem. 1993;208:171–175. - PubMed
- Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. - PubMed
- Fuller CW, Middendorf LR, Benner SA, Church GM, Harris T, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013–1023. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources