A study of the influence of sex on genome wide methylation - PubMed (original) (raw)
A study of the influence of sex on genome wide methylation
Jingyu Liu et al. PLoS One. 2010.
Abstract
Sex differences in methylation status have been observed in specific gene-disease studies and healthy methylation variation studies, but little work has been done to study the impact of sex on methylation at the genome wide locus-to-locus level or to determine methods for accounting for sex in genomic association studies. In this study we investigate the genomic sex effect on saliva DNA methylation of 197 subjects (54 females) using 20,493 CpG sites. Three methods, two-sample T-test, principle component analysis and independent component analysis, all successfully identify sex influences. The results show that sex not only influences the methylation of genes in the X chromosome but also in autosomes. 580 autosomal sites show strong differences between males and females. They are found to be highly involved in eight functional groups, including DNA transcription, RNA splicing, membrane, etc. Equally important is that we identify some methylation sites associated with not only sex, but also other phenotypes (age, smoking and drinking level, and cancer). Verification was done through an independent blood cell DNA methylation data (1298 CpG sites from a cancer panel array). The same genomic site-specific influence pattern and potential confounding effects with cancer were observed. The overlapping rate of identified sex affected genes between saliva and blood cell is 81% for X chromosome, and 8% for autosomes. Therefore, correction for sex is necessary. We propose a simple correction method based on independent component analysis, which is a data driven method and accommodates sample differences. Comparison before and after the correction suggests that the method is able to effectively remove the potentially confounding effects of sex, and leave other phenotypes untouched. As such, our method is able to disentangle the sex influence on a genome wide level, and paves the way to achieve more accurate association analyses in genome wide methylation studies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
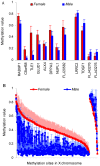
Figure 1. Significant sex effects on 690 methylation sites with mean and standard deviation values.
a): mean and standard deviation methylation values from 12 autosomal sites. Red indicates females and blue indicates males. Bars present mean value, while lines show standard deviation. Eight sites are more methylated in females than males and four sites are more methylated in males than females. b): methylation pattern of 678 sites in X chromosome. They are sorted by female methylation level, presenting 614 sites with higher methylation in females and 64 sites with higher methylation in males. Solid squares show mean value while dash lines show standard deviation.
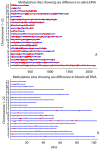
Figure 2. Autosomal sites identified as sex differentially methylated.
Human chromosomes 1-22 are arranged vertically. Methylation sites in each chromosome are plotted horizontally in blue line. Red dots present sites showing sex difference based 5% uncorrected false positive rate. The ones above blue lines are sites where females are more methylated than males. The ones below blue lines are sites where males are more methylated than females. a): result from saliva methylation data. 307 sites are methylated more in females than in males, while 273 sites are methylated more in males. b): results from blood cell verification data. 21 sites are methylated more in females than in males, while 15 sites are methylated more in males.
Figure 3. Weights of the sex-related factor expressed in subjects.
Weights in 143 males are plotted on the left with two subjects above zeros; weights in 54 females are plotted in the right with four subjects below zero.
Similar articles
- An epigenome-wide association study of sex-specific chronological ageing.
McCartney DL, Zhang F, Hillary RF, Zhang Q, Stevenson AJ, Walker RM, Bermingham ML, Boutin T, Morris SW, Campbell A, Murray AD, Whalley HC, Porteous DJ, Hayward C, Evans KL, Chandra T, Deary IJ, McIntosh AM, Yang J, Visscher PM, McRae AF, Marioni RE. McCartney DL, et al. Genome Med. 2019 Dec 31;12(1):1. doi: 10.1186/s13073-019-0693-z. Genome Med. 2019. PMID: 31892350 Free PMC article. - Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets.
Hall E, Volkov P, Dayeh T, Esguerra JL, Salö S, Eliasson L, Rönn T, Bacos K, Ling C. Hall E, et al. Genome Biol. 2014 Dec 3;15(12):522. doi: 10.1186/s13059-014-0522-z. Genome Biol. 2014. PMID: 25517766 Free PMC article. - Sex Differences in the Methylome and Transcriptome of the Human Liver and Circulating HDL-Cholesterol Levels.
García-Calzón S, Perfilyev A, de Mello VD, Pihlajamäki J, Ling C. García-Calzón S, et al. J Clin Endocrinol Metab. 2018 Dec 1;103(12):4395-4408. doi: 10.1210/jc.2018-00423. J Clin Endocrinol Metab. 2018. PMID: 29846646 Free PMC article. - Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis.
Bos SD, Page CM, Andreassen BK, Elboudwarej E, Gustavsen MW, Briggs F, Quach H, Leikfoss IS, Bjølgerud A, Berge T, Harbo HF, Barcellos LF. Bos SD, et al. PLoS One. 2015 Mar 3;10(3):e0117403. doi: 10.1371/journal.pone.0117403. eCollection 2015. PLoS One. 2015. PMID: 25734800 Free PMC article. - Investigating the relationship of DNA methylation with mutation rate and allele frequency in the human genome.
Xia J, Han L, Zhao Z. Xia J, et al. BMC Genomics. 2012;13 Suppl 8(Suppl 8):S7. doi: 10.1186/1471-2164-13-S8-S7. Epub 2012 Dec 17. BMC Genomics. 2012. PMID: 23281708 Free PMC article.
Cited by
- Sex- and tissue-specific methylome changes in brains of mice perinatally exposed to lead.
Sánchez-Martín FJ, Lindquist DM, Landero-Figueroa J, Zhang X, Chen J, Cecil KM, Medvedovic M, Puga A. Sánchez-Martín FJ, et al. Neurotoxicology. 2015 Jan;46:92-100. doi: 10.1016/j.neuro.2014.12.004. Epub 2014 Dec 18. Neurotoxicology. 2015. PMID: 25530354 Free PMC article. - Not all biofluids are created equal: chewing over salivary diagnostics and the epigenome.
Wren ME, Shirtcliff EA, Drury SS. Wren ME, et al. Clin Ther. 2015 Mar 1;37(3):529-39. doi: 10.1016/j.clinthera.2015.02.022. Epub 2015 Mar 13. Clin Ther. 2015. PMID: 25778408 Free PMC article. Review. - miRMap: Profiling 14q32 microRNA Expression and DNA Methylation Throughout the Human Vasculature.
Goossens EAC, de Vries MR, Simons KH, Putter H, Quax PHA, Nossent AY. Goossens EAC, et al. Front Cardiovasc Med. 2019 Aug 8;6:113. doi: 10.3389/fcvm.2019.00113. eCollection 2019. Front Cardiovasc Med. 2019. PMID: 31440517 Free PMC article. - Advances in genome-wide DNA methylation analysis.
Gupta R, Nagarajan A, Wajapeyee N. Gupta R, et al. Biotechniques. 2010 Oct;49(4):iii-xi. doi: 10.2144/000113493. Biotechniques. 2010. PMID: 20964631 Free PMC article. Review. - Sex differences in the intergenerational link between maternal and neonatal whole blood DNA methylation: a genome-wide analysis in 2 birth cohorts.
Hu J, Xu X, Li J, Jiang Y, Hong X, Rexrode KM, Wang G, Hu FB, Zhang H, Karmaus WJ, Wang X, Liang L. Hu J, et al. Clin Epigenetics. 2023 Mar 25;15(1):51. doi: 10.1186/s13148-023-01442-8. Clin Epigenetics. 2023. PMID: 36966332 Free PMC article.
References
- Tost J. Tost J, editor. DNA Methylation: An Introduction to the Biology and the Disease-Associated Changes of a Promising Biomarker. DNA Methylation: Methods and Protocols. 2 ed. 2008. pp. 3–20. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 EB005846/EB/NIBIB NIH HHS/United States
- R01EB005846/EB/NIBIB NIH HHS/United States
- R01 EB006841/EB/NIBIB NIH HHS/United States
- R01 AA012238/AA/NIAAA NIH HHS/United States
- R01AA012238/AA/NIAAA NIH HHS/United States
- R01 EB020407/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials