Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps - PubMed (original) (raw)
Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps
Isheng J Tsai et al. Genome Biol. 2010.
Abstract
Advances in sequencing technology allow genomes to be sequenced at vastly decreased costs. However, the assembled data frequently are highly fragmented with many gaps. We present a practical approach that uses Illumina sequences to improve draft genome assemblies by aligning sequences against contig ends and performing local assemblies to produce gap-spanning contigs. The continuity of a draft genome can thus be substantially improved, often without the need to generate new data.
Figures
Figure 1
Overview of the IMAGE process. Step one, Illumina reads are aligned against the initial assembly. Step two, Illumina reads that align to contig ends, along with their non-aligning mate adjacent to gaps, are assembled into new contigs, which are subsequently mapped back to the initial assembly. Step three, Illumina reads are aligned against the updated assembly and the whole process is repeated iteratively until the gap is closed.
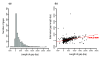
Figure 2
Statistics of sequences at closed gaps in the Echinococcus multilocularis assembly. (a) The frequency of length of newly inserted sequences at gaps. (b) The closed gap length is positively correlated with estimated gap length from the Arachne assembler (Pearson's r = 0.44, P < 0.001).
Figure 3
An example of a gap closed with two iterations of IMAGE in Plasmodium berghei. In the first iteration, IMAGE extended the contig consensus sequence from the right side of the gap, indicated by the green bar. In the second iteration, reads were aligned to the updated contig end. Local assembly of these reads along with their unaligned mates resulted in a new contig to completely close the gap, indicated by the red bar. The horizontal lines above the bars denote the Illumina reads realigned to the updated consensus sequence after each iteration. Below, a zoomed in plot shows the Illumina reads realigned against the closed gap.
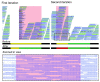
Figure 4
Closing gaps in de novo assembly comprising only Illumina reads. Schematic diagram showing the comparison of the original velvet assembly (3 contigs a, b and c) and the improved assembly in Salmonella enterica. The improved assembly was aligned to the reference sequence with 99.8% identity. The two closed gaps shown were 100% identical to the reference sequence. Contigs are indicated by grey bars; gene annotations are indicated by yellow boxes. Vertical lines highlight the gaps that are filled by IMAGE in the improved contigs. Below, a coverage plot showing the relatively even depth of coverage of realigned Illumina reads at the improved assembly, indicating no signature of misassembly.
Similar articles
- GAPPadder: a sensitive approach for closing gaps on draft genomes with short sequence reads.
Chu C, Li X, Wu Y. Chu C, et al. BMC Genomics. 2019 Jun 6;20(Suppl 5):426. doi: 10.1186/s12864-019-5703-4. BMC Genomics. 2019. PMID: 31167639 Free PMC article. - An improved assembly of the loblolly pine mega-genome using long-read single-molecule sequencing.
Zimin AV, Stevens KA, Crepeau MW, Puiu D, Wegrzyn JL, Yorke JA, Langley CH, Neale DB, Salzberg SL. Zimin AV, et al. Gigascience. 2017 Jan 1;6(1):1-4. doi: 10.1093/gigascience/giw016. Gigascience. 2017. PMID: 28369353 Free PMC article. - High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies.
Aury JM, Cruaud C, Barbe V, Rogier O, Mangenot S, Samson G, Poulain J, Anthouard V, Scarpelli C, Artiguenave F, Wincker P. Aury JM, et al. BMC Genomics. 2008 Dec 16;9:603. doi: 10.1186/1471-2164-9-603. BMC Genomics. 2008. PMID: 19087275 Free PMC article. - De novo assembly of short sequence reads.
Paszkiewicz K, Studholme DJ. Paszkiewicz K, et al. Brief Bioinform. 2010 Sep;11(5):457-72. doi: 10.1093/bib/bbq020. Epub 2010 Aug 19. Brief Bioinform. 2010. PMID: 20724458 Review. - One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly.
Koren S, Phillippy AM. Koren S, et al. Curr Opin Microbiol. 2015 Feb;23:110-20. doi: 10.1016/j.mib.2014.11.014. Epub 2014 Dec 1. Curr Opin Microbiol. 2015. PMID: 25461581 Review.
Cited by
- MetLab: An In Silico Experimental Design, Simulation and Analysis Tool for Viral Metagenomics Studies.
Norling M, Karlsson-Lindsjö OE, Gourlé H, Bongcam-Rudloff E, Hayer J. Norling M, et al. PLoS One. 2016 Aug 1;11(8):e0160334. doi: 10.1371/journal.pone.0160334. eCollection 2016. PLoS One. 2016. PMID: 27479078 Free PMC article. - Defining the phylogenomics of Shigella species: a pathway to diagnostics.
Sahl JW, Morris CR, Emberger J, Fraser CM, Ochieng JB, Juma J, Fields B, Breiman RF, Gilmour M, Nataro JP, Rasko DA. Sahl JW, et al. J Clin Microbiol. 2015 Mar;53(3):951-60. doi: 10.1128/JCM.03527-14. Epub 2015 Jan 14. J Clin Microbiol. 2015. PMID: 25588655 Free PMC article. - GRAbB: Selective Assembly of Genomic Regions, a New Niche for Genomic Research.
Brankovics B, Zhang H, van Diepeningen AD, van der Lee TA, Waalwijk C, de Hoog GS. Brankovics B, et al. PLoS Comput Biol. 2016 Jun 16;12(6):e1004753. doi: 10.1371/journal.pcbi.1004753. eCollection 2016 Jun. PLoS Comput Biol. 2016. PMID: 27308864 Free PMC article. - Comparative genomics of Brachyspira pilosicoli strains: genome rearrangements, reductions and correlation of genetic compliment with phenotypic diversity.
Mappley LJ, Black ML, AbuOun M, Darby AC, Woodward MJ, Parkhill J, Turner AK, Bellgard MI, La T, Phillips ND, La Ragione RM, Hampson DJ. Mappley LJ, et al. BMC Genomics. 2012 Sep 5;13:454. doi: 10.1186/1471-2164-13-454. BMC Genomics. 2012. PMID: 22947175 Free PMC article. - Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of 'Haemophilus intermedius'.
Harris TM, Price EP, Sarovich DS, Nørskov-Lauritsen N, Beissbarth J, Chang AB, Smith-Vaughan HC. Harris TM, et al. Microb Genom. 2020 Jan;6(1):e000303. doi: 10.1099/mgen.0.000303. Microb Genom. 2020. PMID: 31860436 Free PMC article.
References
- Maccallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A, Malek J, McKernan K, Ranade S, Shea TP, Williams L, Young S, Nusbaum C, Jaffe DB. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10:R103. doi: 10.1186/gb-2009-10-10-r103. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous