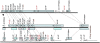Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements - PubMed (original) (raw)
. 2010 Apr 27;107(17):7680-5.
doi: 10.1073/pnas.0910413107. Epub 2010 Apr 13.
H Sezutsu, F Legeai, E Permal, S Bernard-Samain, S Gimenez, C Gagneur, F Cousserans, M Shimomura, A Brun-Barale, T Flutre, A Couloux, P East, K Gordon, K Mita, H Quesneville, P Fournier, R Feyereisen
Affiliations
- PMID: 20388903
- PMCID: PMC2867904
- DOI: 10.1073/pnas.0910413107
Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements
E d'Alençon et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The recent assembly of the silkworm Bombyx mori genome with 432 Mb on 28 holocentric chromosomes has become a reference in the genomic analysis of the very diverse Order of Lepidoptera. We sequenced BACs from two major pests, the noctuid moths Helicoverpa armigera and Spodoptera frugiperda, corresponding to 15 regions distributed on 11 B. mori chromosomes, each BAC/region being anchored by known orthologous gene(s) to analyze syntenic relationships and genome rearrangements among the three species. Nearly 300 genes and numerous transposable elements were identified, with long interspersed nuclear elements and terminal inverted repeats the most abundant transposable element classes. There was a high degree of synteny conservation between B. mori and the two noctuid species. Conserved syntenic blocks of identified genes were very small, however, approximately 1.3 genes per block between B. mori and the two noctuid species and 2.0 genes per block between S. frugiperda and H. armigera. This corresponds to approximately two chromosome breaks per Mb DNA per My. This is a much higher evolution rate than among species of the Drosophila genus and may be related to the holocentric nature of the lepidopteran genomes. We report a large cluster of eight members of the aminopeptidase N gene family that we estimate to have been present since the Jurassic. In contrast, several clusters of cytochrome P450 genes showed multiple lineage-specific duplication events, in particular in the lepidopteran CYP9A subfamily. Our study highlights the value of the silkworm genome as a reference in lepidopteran comparative genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Syntenic relationship at the CYP332A locus. Schemes at scale representing, from top to bottom, H. armigera, S. frugiperda, and B. mori. Arrows represent genes predicted by KAIKOGAAS. Only valid genes are shown (
Fig. S1
). Synteny links are shown with black dotted lines. In text boxes, gene ID, or for other genes, HP, unknown protein (UP; presence of a match to an EST with a threshold of 10−40 by BlastN). Synteny blocks (as defined in text) are shown as colored boxes spanning genes arrows in the case of H. armigera and S. frugiperda, below and above gene arrows in the case of S. frugiperda and B. mori, respectively. See
Fig. S2
for gene name abbreviations.
Fig. 2.
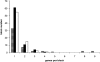
Number of genes per synteny block. Black and white striped bars: between H. armigera and S. frugiperda (average size 2.04). Black bars: between B. mori and S. frugiperda (average size 1.29). White bars: between B. mori and H. armigera (average size 1.32).
Fig. 3.
Correlation between synteny breaks and TE copy density. TE copy density is shown as a function of BAC length. 3A, S. frugiperda CYP4M region; 3B, H. armigera EcR region; dotted lines, TE copy density. Thick lines, regions corresponding to synteny breaks; thin lines. gene density.
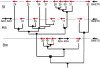
Fig. 4.
Evolution of the CYP9A gene cluster. The CYP9A genes in the three species are shown in their correct orientation and order, but not relative distance, on the BACs or scaffold. From top to bottom: Sf, S. frugiperda; Ha, H. armigera; Bm, B. mori. The phylogenetic tree based on alignments of the CYP proteins is superimposed with its correct topology, but with branch lengths modified for clarity of the figure. Gene duplication events (●), B. mori/noctuid split (i.e., ancient speciation rather than duplication event) (■), and the S. frugiperda/H. armigera split (▲) are indicated. The sequence of events for the CYP9A15, -26, and -27 genes is unresolved. The relative orientation of the genes indicates at least two inversions in addition to the duplication events. Recognized flanking genes [alcohol dehydrogenases (ADH) and BmETS transcription factor] are shown (see
Fig. S2
for details).
Similar articles
- Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping.
Yasukochi Y, Tanaka-Okuyama M, Shibata F, Yoshido A, Marec F, Wu C, Zhang H, Goldsmith MR, Sahara K. Yasukochi Y, et al. PLoS One. 2009 Oct 15;4(10):e7465. doi: 10.1371/journal.pone.0007465. PLoS One. 2009. PMID: 19829706 Free PMC article. - FISH identification of Helicoverpa armigera and Mamestra brassicae chromosomes by BAC and fosmid probes.
Sahara K, Yoshido A, Shibata F, Fujikawa-Kojima N, Okabe T, Tanaka-Okuyama M, Yasukochi Y. Sahara K, et al. Insect Biochem Mol Biol. 2013 Aug;43(8):644-53. doi: 10.1016/j.ibmb.2013.04.003. Epub 2013 Apr 28. Insect Biochem Mol Biol. 2013. PMID: 23628856 - Genomic sequence around butterfly wing development genes: annotation and comparative analysis.
Conceição IC, Long AD, Gruber JD, Beldade P. Conceição IC, et al. PLoS One. 2011;6(8):e23778. doi: 10.1371/journal.pone.0023778. Epub 2011 Aug 31. PLoS One. 2011. PMID: 21909358 Free PMC article. - Comparative study on synteny between yeasts and vertebrates.
Drillon G, Fischer G. Drillon G, et al. C R Biol. 2011 Aug-Sep;334(8-9):629-38. doi: 10.1016/j.crvi.2011.05.011. Epub 2011 Jul 5. C R Biol. 2011. PMID: 21819944 Review. - The Modern View of B Chromosomes Under the Impact of High Scale Omics Analyses.
Ahmad SF, Martins C. Ahmad SF, et al. Cells. 2019 Feb 13;8(2):156. doi: 10.3390/cells8020156. Cells. 2019. PMID: 30781835 Free PMC article. Review.
Cited by
- Holocentromere identity: from the typical mitotic linear structure to the great plasticity of meiotic holocentromeres.
Marques A, Pedrosa-Harand A. Marques A, et al. Chromosoma. 2016 Sep;125(4):669-81. doi: 10.1007/s00412-016-0612-7. Epub 2016 Aug 16. Chromosoma. 2016. PMID: 27530342 Review. - Butterfly genome reveals promiscuous exchange of mimicry adaptations among species.
Heliconius Genome Consortium. Heliconius Genome Consortium. Nature. 2012 Jul 5;487(7405):94-8. doi: 10.1038/nature11041. Nature. 2012. PMID: 22722851 Free PMC article. - Advances and Challenges of Using the Sterile Insect Technique for the Management of Pest Lepidoptera.
Marec F, Vreysen MJB. Marec F, et al. Insects. 2019 Oct 25;10(11):371. doi: 10.3390/insects10110371. Insects. 2019. PMID: 31731445 Free PMC article. Review. - Gypsy moth genome provides insights into flight capability and virus-host interactions.
Zhang J, Cong Q, Rex EA, Hallwachs W, Janzen DH, Grishin NV, Gammon DB. Zhang J, et al. Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1669-1678. doi: 10.1073/pnas.1818283116. Epub 2019 Jan 14. Proc Natl Acad Sci U S A. 2019. PMID: 30642971 Free PMC article. - Isolation of BAC clones containing conserved genes from libraries of three distantly related moths: a useful resource for comparative genomics of Lepidoptera.
Yasukochi Y, Tanaka-Okuyama M, Kamimura M, Nakano R, Naito Y, Ishikawa Y, Sahara K. Yasukochi Y, et al. J Biomed Biotechnol. 2011;2011:165894. doi: 10.1155/2011/165894. Epub 2010 Nov 25. J Biomed Biotechnol. 2011. PMID: 21127704 Free PMC article.
References
- The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. - PubMed
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. - PubMed
- Gall LF, Tiffney BH. A Fossil Noctuid Moth Egg from the Late Cretaceous of Eastern North America. Science. 1983;219:507–509. - PubMed
- Grimaldi D, Engel MS. Evolution of the Insects. Cambridge: Cambridge University Press; 2005. p. 755.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous