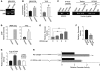HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype - PubMed (original) (raw)
. 2010 May;120(5):1535-50.
doi: 10.1172/JCI39534. Epub 2010 Apr 12.
Janna K Mouw, Meredith A Unger, Johnathon N Lakins, Mawuse K Gbegnon, Virginia B Clemmer, Miriam Benezra, Jonathan D Licht, Nancy J Boudreau, Kelvin K C Tsai, Alana L Welm, Michael D Feldman, Barbara L Weber, Valerie M Weaver
Affiliations
- PMID: 20389018
- PMCID: PMC2860938
- DOI: 10.1172/JCI39534
HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype
Penney M Gilbert et al. J Clin Invest. 2010 May.
Abstract
Breast cancer 1, early onset (BRCA1) expression is often reduced in sporadic breast tumors, even in the absence of BRCA1 genetic modifications, but the molecular basis for this is unknown. In this study, we identified homeobox A9 (HOXA9) as a gene frequently downregulated in human breast cancers and tumor cell lines and noted that reduced HOXA9 transcript levels associated with tumor aggression, metastasis, and patient mortality. Experiments revealed that loss of HOXA9 promoted mammary epithelial cell growth and survival and perturbed tissue morphogenesis. Restoring HOXA9 expression repressed growth and survival and inhibited the malignant phenotype of breast cancer cells in culture and in a xenograft mouse model. Molecular studies showed that HOXA9 restricted breast tumor behavior by directly modulating the expression of BRCA1. Indeed, ectopic expression of wild-type BRCA1 phenocopied the tumor suppressor function of HOXA9, and reducing BRCA1 levels or function inhibited the antitumor activity of HOXA9. Consistently, HOXA9 expression correlated with BRCA1 in clinical specimens and with tumor aggression in patients lacking estrogen receptor/progesterone receptor expression in their breast tissue. These findings indicate that HOXA9 restricts breast tumor aggression by modulating expression of the tumor suppressor gene BRCA1, which we believe provides an explanation for the loss of BRCA1 expression in sporadic breast tumors in the absence of BRCA1 genetic modifications.
Figures
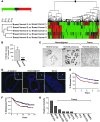
Figure 1. Breast malignancy is associated with reduced HOXA9 expression.
(A) Cluster diagram of Affymetrix microarray data using Rosetta Resolver to compare gene expression profiles of matched normal mammary tissue and adjacent primary breast cancers, revealing significantly lower HOXA9 transcript levels in 4 out of 5 expression sets analyzed (P ≤ 0.01). (B) Levels of HOXA9 mRNA (by quantitative RT-PCR) in primary human mammary tumors (n = 47) compared with normal breast tissue (n = 16). ****P = 0.00035. (C) In situ hybridization using a HOXA9 probe on nonmalignant (n = 4) or malignant (n = 6) mammary epithelial tissue. Scale bar: 100 μm. (D) Immunofluorescent staining for HOXA9 (red) and nuclei (blue) demonstrates robust cytoplasmic and nuclear localized HOXA9 protein in the epithelium of nonmalignant human breast tissue and reduced levels in breast tumors. Top right insets (original magnification, ×20) show a broader view of the breast tissue, with arrows indicating regions blown up in the main images. The arrow in the center main image corresponds with the bottom left inset, which shows a view of HOXA9 staining (original magnification, ×30). Scale bar: 50 mm. The 2o control shows no nonspecific staining. (E) Breast cancer patients whose tumors expressed the lowest HOXA9 level (lowest expression quartile; red line) experienced significantly reduced disease-free survival compared with all other patients in the study (blue line). An “X” is used to denote each censored sample. P = 0.025. (F) Patients with the lowest HOXA9 levels in their tumors (lowest expression quartile; red line) also had significantly increased metastasis as a first event when compared with all other patients (blue line). An “X” is used to denote each censored sample. P = 0.02. (G) Bar graph demonstrating relative HOXA9 gene expression levels in nonmalignant and malignant MEC lines.
Figure 2. HOXA9 modulates the growth and survival of breast cancer cells.
(A) Semiquantitative PCR gel, indicating HOXA9 mRNA levels expressed in human nonmalignant (MCF10A), metastatic (MDA-231), and transformed (T4-2) MECs. 18S rRNA was used as a control. (B) Semiquantitative PCR gel showing transgenic HOXA9 mRNA levels in MDA-231 and T4-2 cell lines. Immunofluorescence images of nuclei (blue) and FLAG-tagged HOXA9 (red) in MDA-231 and T4-2 cells. Arrows indicate localization of Flag-tagged HOXA9-positive cells. Scale bar: 10 μm. (C) Proliferation in MDA 231 and T4-2 cells following HOXA9 reexpression. **P = 0.0025, ***P = 0.0003. (D) Cross-sectional area of MDA-231 and T4-2 breast tumor colonies in rBM expressing either the vector or HOXA9 transgene. ****P = 0.0001, **P = 0.0068. (E) Immunofluorescence images of β4 integrin (red), Laminin-5 (red), β-catenin (red), and nuclei (blue) in T4-2 colonies expressing the vector or HOXA9 or phenotypically reverted acini (anti-EGFR) by inhibiting EGFR activity using tyrphostin. Arrows indicate cleared lumen. Scale bar: 10 μm. (F) Lumens observed in rBM-generated T4-2 colonies expressing the vector, HOXA9, or anti-EGFR phenotypically reverted acini. *P = 0.0188, **P = 0.0076. (G) The percentage of tumor colonies greater than 30 μm in diameter, and phase contrast images of tumor colonies embedded within soft agar. Scale bar: 50 μm (top panel); 20 μm (bottom panel). *P = 0.0221, **P = 0.018, ***P = 0.0005. (H) High- and low-magnification phase images of H&E-stained tissue sections of control tumor (T4-2 vector) and HOXA9 reexpressing tumor (T4-2 HOXA9) xenografts. Vascularized regions of T4-2 control tumors are indicated with white arrows, and cystic regions of T4-2 HoxA9 reexpressing tumors are indicate with black arrows. Original magnification, ×40; ×10 (insets). Scale bar: 100 μm.
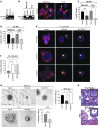
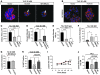
Figure 3. HOXA9 regulates BRCA1 expression.
(A) Semiquantitative PCR gel, indicating increased BRCA1 expression with the reexpression of HOXA9 in MDA-231 cells. (B) Bar graph quantifying immunoblot data from multiple experiments, showing increased BRCA1 protein in MDA-231 or T4-2 breast tumor cells reexpressing HOXA9. *P = 0.0457, **P = 0.0028. (C) Representative gel of ChIP studies in breast cancer cells, revealing coprecipitation of HOXA9 with the BRCA1 promoter and acetylated acetyl-H3-histone with the β-globin promoter. (D) Bar graphs quantifying ChIP experiments in MDA-231 (n = 2) and T4-2 cells (*P = 0.0178; n = 4). (E) Luciferase reporter analysis, showing a dose-dependent increase in BRCA1 promoter activity in response to addition of wild-type HOXA9. **P = 0.001. (F) Luciferase reporter analysis, displaying loss of BRCA1 promoter activity upon addition of HOXA9 containing an N255T (DNA BM) mutation in the conserved DNA binding domain. *P = 0.03 (G) Luciferase reporter analysis, indicating enhanced HOXA9-mediated BRCA1 promoter activity upon addition of PBX1 cofactor (2 μg), compared with Pbx1 alone or Pbx1 cotransfected with a shRNA reducing HOXA9 expression. *P = 0.0259. (H) Luciferase reporter analysis, showing a diminished responsiveness of a BRCA1 promoter construct containing a deletion in residues –223 to +44, which contains 3 putative Hox binding sites (gray bar). Data are normalized to matched vector control (black bars). Negative numbers refer to basepairs upstream of the BRCA1 transcription start site. **P = 0.02.
Figure 4. HOXA9 regulates nonmalignant MEC growth by modulating BRCA1 expression.
(A) Immunofluorescence images of β-catenin (red), β4 integrin (green), Laminin-5 (green), and nuclei (blue) in MCF-10A colonies expressing either luciferase control shRNA or shRNA-HOXA9 clone 3. Arrows indicate cleared lumen. Scale bar: 50 μm. (B) Colony size of MCF-10A cells cultured within a rBM and expressing reduced HOXA9 levels. ****P = 0.0001. (C) Lumens observed in MCF-10A colonies expressing luciferase control shRNA as compared with those with shRNA-mediated HOXA9 knockdown. **P = 0.0010. (D) The response (BRCA1 protein levels) of MCF-10A cells expressing reduced HOXA9 levels to 5 Gray irradiation compared with nonirradiated samples. *P < 0.001. (E and F) Phase images of tumor colonies embedded within soft agar (E) and the percentage of colonies greater than 40 μm in diameter (F). Scale bar: 50 μm. *P = 0.001. (G) Immunofluorescence images of β-catenin (red) and nuclei (blue) in MCF-10A colonies, expressing either luciferase control shRNA or shRNA-BRCA1 clone 5. The arrow indicates cleared lumen. Scale bar: 50 μm. (H) Colony size of MCF-10A cells cultured within a rBM and expressing reduced BRCA1 levels. ***P = 0.0024. (I) Quantification of lumens observed in MCF-10A colonies expressing luciferase control shRNA as compared with those with shRNA-mediated BRCA1 knockdown. ****P = 0.0003. (J) Immunofluorescence images of β-catenin (red) and nuclei (blue) in MCF-10A colonies expressing vector or mutant BRCA1 (BRCA1 MT). The arrow indicates cleared lumen. Scale bar: 10 μm. (K) Quantification of cross-sectional area of MCF-10A colonies in cells cultured within a rBM and coexpressing mutant BRCA1. *P = 0.05. (L) Quantification of lumens observed in MCF-10A colonies expressing vector control as compared with those expressing a mutant BRCA1. *P = 0.05.
Figure 5. HOXA9 regulates BRCA1 to repress the malignant behavior of MECs.
(A) Immunofluorescence images of β-catenin (red) and nuclei (blue) in T4-2 colonies reexpressing HOXA9 or BRCA1. Arrows indicate cleared lumens. Scale bar: 10 μm. (B) Immunofluorescence images of β-catenin (red) and nuclei (blue) in T4-2 colonies reexpressing HOXA9 alone or with a mutant BRCA1. Scale bar: 10 μm. (C) Quantification of colony size of T4-2 cells reexpressing HOXA9 or BRCA1. ***P = 0.001. (D) Quantification of cross-sectional area of T4-2 colonies formed by cells reexpressing HOXA9 alone or with a mutant BRCA1. *P = 0.05, ***P = 0.001. (E) Quantification of anchorage-independent growth and survival in T4-2 cells reexpressing either HOXA9 or BRCA1. ***P = 0.001. (F) Quantification of the percentage of T4-2 colonies that formed lumens following the reexpression of HOXA9 or BRCA1. *P = 0.0263. (G) Quantification of lumen formation in rBM T4-2 colonies with reexpressed HOXA9, when BRCA1 function has been compromised through coexpression of a mutant BRCA1. (H) Quantification of proliferation in T4-2 cells following HOXA9 reexpression with a mutant BRCA1. **P = 0.0025, ***P = 0.0003. (I) The time course of the progressive increase in xenograft size (5–30 days). Reexpression of HOXA9 in T4-2 tumor cells significantly reduced the rate of lesion expansion (filled circles) compared with the T4-2 vector controls (filled squares), which could be restored to that of T4-2 breast tumor cells if coexpressed with a mutant BRCA1 (filled triangles with red line). ***P = 0.001. (J) Lesion size (28 days) in each experimental group. **P = 0.01, ***P = 0.001.
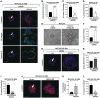
Figure 6. Clinical correlation between HOXA9 and BRCA1 expression.
(A) Line graph illustrating that significant correlations exist between HOXA9 and BRCA1 mRNA levels expressed in a cohort of normal (shown in blue; P ≤ 0.0001; _r_2 = 0.8534) and tumorigenic (shown in red; P ≤ 0.0001; _r_2 = 0.7160) human breast tissue specimens (n = 53). (B) Graph illustrating the relationship between HOXA9 and BRCA1 protein levels in human ER–/PR–/ErbB2– breast samples. (C) Immunohistochemistry showing colocalized expression of HOXA9 and BRCA1 protein in the epithelium of normal human breast tissue and HOXA9+/BRCA1+ tumor samples, as compared with HOXA9–/BRCA1– tumor samples. (HOXA9 was “stained” with Fast Red, and BRCA1 was “stained” with DAB). Scale bar: 100 μm.
Comment in
- Tumour suppression: Double act.
McCarthy N. McCarthy N. Nat Rev Cancer. 2010 Jun;10(6):382. doi: 10.1038/nrc2858. Nat Rev Cancer. 2010. PMID: 20506586 No abstract available.
Similar articles
- Loss of nuclear BRCA1 expression in breast cancers is associated with a highly proliferative tumor phenotype.
Jarvis EM, Kirk JA, Clarke CL. Jarvis EM, et al. Cancer Genet Cytogenet. 1998 Mar;101(2):109-15. doi: 10.1016/s0165-4608(97)00267-7. Cancer Genet Cytogenet. 1998. PMID: 9494611 - Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss in BRCA1-associated cases.
Yoshikawa K, Honda K, Inamoto T, Shinohara H, Yamauchi A, Suga K, Okuyama T, Shimada T, Kodama H, Noguchi S, Gazdar AF, Yamaoka Y, Takahashi R. Yoshikawa K, et al. Clin Cancer Res. 1999 Jun;5(6):1249-61. Clin Cancer Res. 1999. PMID: 10389907 - TNRC9 downregulates BRCA1 expression and promotes breast cancer aggressiveness.
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK, Ascierto ML, Sastry KS, Bedri S, Kizhakayil D, Aigha II, Malek J, Al-Bozom I, Gehani S, Furtado S, Mathiowitz E, Wang E, Marincola FM, Chouchane L. Shan J, et al. Cancer Res. 2013 May 1;73(9):2840-9. doi: 10.1158/0008-5472.CAN-12-4313. Epub 2013 Feb 27. Cancer Res. 2013. PMID: 23447579 - Promotion of BRCA1-associated triple-negative breast cancer by ovarian hormones.
Lee EY. Lee EY. Curr Opin Obstet Gynecol. 2008 Feb;20(1):68-73. doi: 10.1097/GCO.0b013e3282f42237. Curr Opin Obstet Gynecol. 2008. PMID: 18197009 Review. - BRACking news on triple-negative/basal-like breast cancers: how BRCA1 deficiency may result in the development of a selective tumor subtype.
Santarosa M, Maestro R. Santarosa M, et al. Cancer Metastasis Rev. 2012 Jun;31(1-2):131-42. doi: 10.1007/s10555-011-9336-6. Cancer Metastasis Rev. 2012. PMID: 22101651 Review.
Cited by
- Analysis of HOX gene expression patterns in human breast cancer.
Hur H, Lee JY, Yun HJ, Park BW, Kim MH. Hur H, et al. Mol Biotechnol. 2014 Jan;56(1):64-71. doi: 10.1007/s12033-013-9682-4. Mol Biotechnol. 2014. PMID: 23820980 - Growth and differentiation factor 3 induces expression of genes related to differentiation in a model of cancer stem cells and protects them from retinoic acid-induced apoptosis.
Tykwinska K, Lauster R, Knaus P, Rosowski M. Tykwinska K, et al. PLoS One. 2013 Aug 12;8(8):e70612. doi: 10.1371/journal.pone.0070612. eCollection 2013. PLoS One. 2013. PMID: 23950971 Free PMC article. - Homeobox Genes in Cancers: From Carcinogenesis to Recent Therapeutic Intervention.
Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo X, Huang W, Xia L. Feng Y, et al. Front Oncol. 2021 Oct 14;11:770428. doi: 10.3389/fonc.2021.770428. eCollection 2021. Front Oncol. 2021. PMID: 34722321 Free PMC article. Review. - The Pbx interaction motif of Hoxa1 is essential for its oncogenic activity.
Delval S, Taminiau A, Lamy J, Lallemand C, Gilles C, Noël A, Rezsohazy R. Delval S, et al. PLoS One. 2011;6(9):e25247. doi: 10.1371/journal.pone.0025247. Epub 2011 Sep 21. PLoS One. 2011. PMID: 21957483 Free PMC article. - MNX1 Promotes Anti-HER2 Therapy Sensitivity via Transcriptional Regulation of CD-M6PR in HER2-Positive Breast Cancer.
Chi W, Xiu B, Xiong M, Wang X, Li P, Zhang Q, Hou J, Sang Y, Zhou X, Chen M, Zheng S, Zhang L, Xue J, Chi Y, Wu J. Chi W, et al. Int J Mol Sci. 2023 Dec 22;25(1):221. doi: 10.3390/ijms25010221. Int J Mol Sci. 2023. PMID: 38203393 Free PMC article.
References
- Chen H, Sukumar S. HOX genes: emerging stars in cancer. Cancer Biol Ther. 2003;2(5):524–525. - PubMed
- Nakamura T, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BC062562/BC/NCI NIH HHS/United States
- R01 CA078731/CA/NCI NIH HHS/United States
- U54CA143836/CA/NCI NIH HHS/United States
- RS1-00449/RS/DRS NIH HHS/United States
- A107165/PHS HHS/United States
- U54 CA143836/CA/NCI NIH HHS/United States
- T32 CA009151/CA/NCI NIH HHS/United States
- R01 CA138818/CA/NCI NIH HHS/United States
- R01-CA078731/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous