Development and evaluation of an ompA quantitative real-time PCR assay for Chlamydia trachomatis serovar determination - PubMed (original) (raw)
Development and evaluation of an ompA quantitative real-time PCR assay for Chlamydia trachomatis serovar determination
Matthew P Stevens et al. J Clin Microbiol. 2010 Jun.
Abstract
Knowledge of circulating Chlamydia trachomatis serovars can be beneficial for sexual network surveillance, monitoring treatment success, and associating specific clinical manifestations. Typically, C. trachomatis serovars are predicted by nucleotide sequencing of four variable domains within the ompA gene. However, sequencing procedures can be labor-intensive, are not readily available, and can lack the capacity to identify multiple serovars. This study describes the development and evaluation of a quantitative real-time PCR (qPCR) test algorithm for the rapid prediction of C. trachomatis serovars, including ocular (A to C) and anogenital (D to L3) strains. This test comprises a primary qPCR to confirm C. trachomatis positivity and the phylogenetic group(s) present and a secondary set of qPCRs to determine specific serovars. Cell culture isolates from 15 prototypic C. trachomatis serovars were correctly identified using this assay, with no cross-reactivity observed among serovars or with other common pathogenic microorganisms. Five hundred clinical specimens (previously diagnosed as being C. trachomatis positive) were evaluated by qPCR, with their results compared to results obtained by conventional sequencing. The qPCR identified 88.9% (423/476) complete matches (95% confidence interval [CI], 86 to 92%) of serovars compared to the results obtained using the sequence-based approach. Among the anogenital specimens, 2.4% (12/494) (95% CI, 1.3 to 4.2%) contained multiple serovars, categorized as single-serovar infections by conventional sequencing. Overall, this test exhibited high discriminatory success for predicting C. trachomatis serovars, particularly among anogenital infections. This is the first report of a qPCR typing assay offering differentiation of C. trachomatis serovars associated with both anogenital and ocular diseases.
Figures
FIG. 1.
Schematic representation of the ompA gene regions amplified by qPCRs for C. trachomatis serovar determination. Primers are shown by left/right arrows, and probes are indicated by serovar letters, with those above annealing to the sense strand and those below annealing to the antisense strand. *, C. trachomatis group-specific probes (primary PCR is shaded).
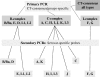
FIG. 2.
Schematic representation of the qPCR testing algorithm used for determining C. trachomatis serovars.
Similar articles
- Rapid detection of Chlamydia trachomatis and typing of the Lymphogranuloma venereum associated L-Serovars by TaqMan PCR.
Schaeffer A, Henrich B. Schaeffer A, et al. BMC Infect Dis. 2008 Apr 30;8:56. doi: 10.1186/1471-2334-8-56. BMC Infect Dis. 2008. PMID: 18447917 Free PMC article. - Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis.
Xiong L, Kong F, Zhou H, Gilbert GL. Xiong L, et al. J Clin Microbiol. 2006 Apr;44(4):1413-8. doi: 10.1128/JCM.44.4.1413-1418.2006. J Clin Microbiol. 2006. PMID: 16597870 Free PMC article. - Combination of PCR targeting the VD2 of omp1 and reverse line blot analysis for typing of urogenital Chlamydia trachomatis serovars in cervical scrape specimens.
Molano M, Meijer CJ, Morré SA, Pol R, van den Brule AJ. Molano M, et al. J Clin Microbiol. 2004 Jul;42(7):2935-9. doi: 10.1128/JCM.42.7.2935-2939.2004. J Clin Microbiol. 2004. PMID: 15243041 Free PMC article. - Molecular typing of Chlamydia trachomatis: An overview.
Rawre J, Juyal D, Dhawan B. Rawre J, et al. Indian J Med Microbiol. 2017 Jan-Mar;35(1):17-26. doi: 10.4103/ijmm.IJMM_16_341. Indian J Med Microbiol. 2017. PMID: 28303813 Review. - Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism.
Abdelsamed H, Peters J, Byrne GI. Abdelsamed H, et al. Future Microbiol. 2013 Sep;8(9):1129-1146. doi: 10.2217/fmb.13.80. Future Microbiol. 2013. PMID: 24020741 Free PMC article. Review.
Cited by
- Chlamydia trachomatis genovar distribution in clinical urogenital specimens from Tunisian patients: high prevalence of C. trachomatis genovar E and mixed infections.
Gharsallah H, Frikha-Gargouri O, Sellami H, Besbes F, Znazen A, Hammami A. Gharsallah H, et al. BMC Infect Dis. 2012 Nov 30;12:333. doi: 10.1186/1471-2334-12-333. BMC Infect Dis. 2012. PMID: 23198910 Free PMC article. - Chlamydia trachomatis incidence and re-infection among young women--behavioural and microbiological characteristics.
Walker J, Tabrizi SN, Fairley CK, Chen MY, Bradshaw CS, Twin J, Taylor N, Donovan B, Kaldor JM, McNamee K, Urban E, Walker S, Currie M, Birden H, Bowden F, Gunn J, Pirotta M, Gurrin L, Harindra V, Garland SM, Hocking JS. Walker J, et al. PLoS One. 2012;7(5):e37778. doi: 10.1371/journal.pone.0037778. Epub 2012 May 25. PLoS One. 2012. PMID: 22662220 Free PMC article. - Mycoplasma genitalium in the Far North Queensland backpacker population: An observational study of prevalence and azithromycin resistance.
Trevis T, Gossé M, Santarossa N, Tabrizi S, Russell D, McBride WJ. Trevis T, et al. PLoS One. 2018 Aug 28;13(8):e0202428. doi: 10.1371/journal.pone.0202428. eCollection 2018. PLoS One. 2018. PMID: 30153259 Free PMC article. - Chlamydia trachomatis genotypes in a cross-sectional study of urogenital samples from remote Northern and Central Australia.
Giffard PM, Brenner NC, Tabrizi SN, Garland SM, Holt DC, Andersson P, Lilliebridge RA, Tong SY, Karimi M, Boylan P, Ryder N, Johns T, Singh G. Giffard PM, et al. BMJ Open. 2016 Jan 6;6(1):e009624. doi: 10.1136/bmjopen-2015-009624. BMJ Open. 2016. PMID: 26739733 Free PMC article. - Combination of compound screening with an animal model identifies pentamidine to prevent Chlamydia trachomatis infection.
Knapp K, Klasinc R, Koren A, Siller M, Dingelmaier-Hovorka R, Drach M, Sanchez J, Chromy D, Kranawetter M, Grimm C, Bergthaler A, Kubicek S, Stockinger H, Stary G. Knapp K, et al. Cell Rep Med. 2024 Jul 16;5(7):101643. doi: 10.1016/j.xcrm.2024.101643. Epub 2024 Jul 8. Cell Rep Med. 2024. PMID: 38981484 Free PMC article.
References
- Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. - PMC - PubMed
- Cates, W., Jr., and J. N. Wasserheit. 1991. Genital chlamydial infections: epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771-1781. - PubMed
- Garland, S. M., and B. Johnson. 1989. Chlamydia trachomatis infections—The Royal Women's Hospital experience. Med. J. Aust. 150:174-177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical