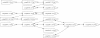Finishing genomes with limited resources: lessons from an ensemble of microbial genomes - PubMed (original) (raw)
Finishing genomes with limited resources: lessons from an ensemble of microbial genomes
Niranjan Nagarajan et al. BMC Genomics. 2010.
Abstract
While new sequencing technologies have ushered in an era where microbial genomes can be easily sequenced, the goal of routinely producing high-quality draft and finished genomes in a cost-effective fashion has still remained elusive. Due to shorter read lengths and limitations in library construction protocols, shotgun sequencing and assembly based on these technologies often results in fragmented assemblies. Correspondingly, while draft assemblies can be obtained in days, finishing can take many months and hence the time and effort can only be justified for high-priority genomes and in large sequencing centers. In this work, we revisit this issue in light of our own experience in producing finished and nearly-finished genomes for a range of microbial species in a small-lab setting. These genomes were finished with surprisingly little investments in terms of time, computational effort and lab work, suggesting that the increased access to sequencing might also eventually lead to a greater proportion of finished genomes from small labs and genomics cores.
Figures
Figure 1
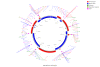
Summary of the finishing effort for A. aphrophilus. As can be seen from the figure much of the finishing effort (PCR experiments indicated by bars in the outermost ring) were devoted to disambiguating the neighborhood of the rRNA operon.
Figure 2
R. prowazekii Contig Graph. Note that the comments for Figure 3 are also valid here. This graph can be resolved into a unique in silico reconstruction of the genome.
Figure 3
Partial Contig Graph of Y. kristensenii. The pointed boxes represent contigs while the edges mark the presence of reads that span the corresponding contigs. The arrows on both ends of an edge indicate the orientation of the adjacent contigs. An arrow "out" of a contig indicates that the end of the contig is adjacent and an arrow "in" indicates that the beginning of the contig is adjacent.
Figure 4
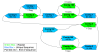
AMOS-Hybrid pipeline. Circles are used to represent input/output and intermediate datasets. Names in parentheses refer to the programs used to perform the corresponding tasks in the boxes.
Figure 5
The optical mapping process. To generate a whole-genome optical map, DNA is sheared into fragments that are stretched and fixed onto an optical mapping surface and then digested using a restriction enzyme. The resulting pieces are optically analyzed and assembled into a genome-wide map.
Similar articles
- Reducing assembly complexity of microbial genomes with single-molecule sequencing.
Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, Mcvey SD, Radune D, Bergman NH, Phillippy AM. Koren S, et al. Genome Biol. 2013;14(9):R101. doi: 10.1186/gb-2013-14-9-r101. Genome Biol. 2013. PMID: 24034426 Free PMC article. - The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation.
Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, Cottingham RW, Kyrpides NC. Mavromatis K, et al. PLoS One. 2012;7(12):e48837. doi: 10.1371/journal.pone.0048837. Epub 2012 Dec 12. PLoS One. 2012. PMID: 23251337 Free PMC article. - Completion of draft bacterial genomes by long-read sequencing of synthetic genomic pools.
Derakhshani H, Bernier SP, Marko VA, Surette MG. Derakhshani H, et al. BMC Genomics. 2020 Jul 29;21(1):519. doi: 10.1186/s12864-020-06910-6. BMC Genomics. 2020. PMID: 32727443 Free PMC article. - Some lessons from Rickettsia genomics.
Renesto P, Ogata H, Audic S, Claverie JM, Raoult D. Renesto P, et al. FEMS Microbiol Rev. 2005 Jan;29(1):99-117. doi: 10.1016/j.femsre.2004.09.002. FEMS Microbiol Rev. 2005. PMID: 15652978 Review. - One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly.
Koren S, Phillippy AM. Koren S, et al. Curr Opin Microbiol. 2015 Feb;23:110-20. doi: 10.1016/j.mib.2014.11.014. Epub 2014 Dec 1. Curr Opin Microbiol. 2015. PMID: 25461581 Review.
Cited by
- BAIT: Organizing genomes and mapping rearrangements in single cells.
Hills M, O'Neill K, Falconer E, Brinkman R, Lansdorp PM. Hills M, et al. Genome Med. 2013 Sep 13;5(9):82. doi: 10.1186/gm486. eCollection 2013. Genome Med. 2013. PMID: 24028793 Free PMC article. - De novo likelihood-based measures for comparing genome assemblies.
Ghodsi M, Hill CM, Astrovskaya I, Lin H, Sommer DD, Koren S, Pop M. Ghodsi M, et al. BMC Res Notes. 2013 Aug 22;6:334. doi: 10.1186/1756-0500-6-334. BMC Res Notes. 2013. PMID: 23965294 Free PMC article. - Enhanced de novo assembly of high throughput pyrosequencing data using whole genome mapping.
Onmus-Leone F, Hang J, Clifford RJ, Yang Y, Riley MC, Kuschner RA, Waterman PE, Lesho EP. Onmus-Leone F, et al. PLoS One. 2013 Apr 17;8(4):e61762. doi: 10.1371/journal.pone.0061762. Print 2013. PLoS One. 2013. PMID: 23613926 Free PMC article. - Approaches for in silico finishing of microbial genome sequences.
Kremer FS, McBride AJA, Pinto LS. Kremer FS, et al. Genet Mol Biol. 2017 Jul-Sep 01;40(3):553-576. doi: 10.1590/1678-4685-GMB-2016-0230. Genet Mol Biol. 2017. PMID: 28898352 Free PMC article. - Genome sequencing of bacteria: sequencing, de novo assembly and rapid analysis using open source tools.
Kisand V, Lettieri T. Kisand V, et al. BMC Genomics. 2013 Apr 1;14:211. doi: 10.1186/1471-2164-14-211. BMC Genomics. 2013. PMID: 23547799 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous