Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma - PubMed (original) (raw)
. 2010 May 18;17(5):510-22.
doi: 10.1016/j.ccr.2010.03.017. Epub 2010 Apr 15.
Daniel J Weisenberger, Kristin Diefes, Heidi S Phillips, Kanan Pujara, Benjamin P Berman, Fei Pan, Christopher E Pelloski, Erik P Sulman, Krishna P Bhat, Roel G W Verhaak, Katherine A Hoadley, D Neil Hayes, Charles M Perou, Heather K Schmidt, Li Ding, Richard K Wilson, David Van Den Berg, Hui Shen, Henrik Bengtsson, Pierre Neuvial, Leslie M Cope, Jonathan Buckley, James G Herman, Stephen B Baylin, Peter W Laird, Kenneth Aldape; Cancer Genome Atlas Research Network
Affiliations
- PMID: 20399149
- PMCID: PMC2872684
- DOI: 10.1016/j.ccr.2010.03.017
Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma
Houtan Noushmehr et al. Cancer Cell. 2010.
Abstract
We have profiled promoter DNA methylation alterations in 272 glioblastoma tumors in the context of The Cancer Genome Atlas (TCGA). We found that a distinct subset of samples displays concerted hypermethylation at a large number of loci, indicating the existence of a glioma-CpG island methylator phenotype (G-CIMP). We validated G-CIMP in a set of non-TCGA glioblastomas and low-grade gliomas. G-CIMP tumors belong to the proneural subgroup, are more prevalent among lower-grade gliomas, display distinct copy-number alterations, and are tightly associated with IDH1 somatic mutations. Patients with G-CIMP tumors are younger at the time of diagnosis and experience significantly improved outcome. These findings identify G-CIMP as a distinct subset of human gliomas on molecular and clinical grounds.
(c) 2010 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest statement
P.W.L. is a shareholder, consultant and scientific advisory board member of Epigenomics, AG, which has a commercial interest in DNA methylation markers. This work was not supported by Epigenomics, AG. K.A. is a consultant and scientific advisory board member for Castle Biosciences, which has a commercial interest in molecular diagnostics. This work was not supported by Castle Biosciences.
Figures
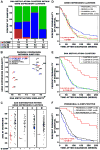
Figure 1. Clustering of TCGA GBM tumors and control samples identifies a CpG Island Methylator Phenotype (G-CIMP)
Unsupervised consensus clustering was performed using the 1,503 Infnium DNA methylation probes whose DNA methylation beta values varied the most across the 91 TCGA GBM samples. DNA Methylation clusters are distinguished with a color code at the top of the panel: red, consensus cluster 1 (n=12 tumors); blue, consensus cluster 2 (n=31 tumors); green, consensus cluster 3 (n=48 samples). Each sample within each DNA Methylation cluster are colored labeled as described in the key for its gene expression cluster membership (Proneural, Neural, Classical and Mesenchymal). The somatic mutation status of five genes (EGFR, IDH1, NF1, PTEN, and TP53) are indicated by the black squares, the gray squares indicate the absence of mutations in the sample and the white squares indicate that the gene was not screened in the specific sample. G-CIMP-positive samples are labeled at the bottom of the matrix. A) Consensus matrix produced by k-means clustering (K = 3). The samples are listed in the same order on the x and y axes. Consensus index values range from 0 to 1, 0 being highly dissimilar and 1 being highly similar. B) One-dimensional hierarchical clustering of the same 1,503 most variant probes, with retention of the same sample order as in Figure 1A. Each row represents a probe; each column represents a sample. The level of DNA methylation (beta value) for each probe, in each sample, is represented by using a color scale as shown in the legend; white indicates missing data. M._Sss_I-treated DNA (n=2), WGA-DNA (n=2) and normal brain (n=4) samples are included in the heatmap but did not contribute to the unsupervised clustering. The probes in the eight control samples are listed in the same order as the y axis of the GBM sample heatmap. See also Figure S1 and Table S1.
Figure 2. Characterization of G-CIMP tumors as a unique subtype of GBMs within the proneural gene expression subgroup
A) Integration of the samples within each DNA methylation and gene expression cluster. Samples are primarily categorized by their gene expression subtype: P, proneural; N, neural; C, classical; M, mesenchymal. The number and percent of tumors within each DNA methylation cluster (red, cluster 1 (G-CIMP); blue, cluster 2; green, cluster 3) are indicated for each gene expression subtype. B) Scatter plot of pairwise comparison of the gene expression and DNA methylation clusters as identified in Figure 1 and Figure S1. Same two-letter represents self-comparison while mixed two-letter represents the pair-wise correlation between gene expression and DNA methylation. Axes are reversed to illustrate increasing similarity. C) GBM patient age distribution at time of diagnosis within each gene expression cluster. Samples are divided by gene expression clusters as identified along the top of each jitter plot, and further subdivided by G-CIMP status within each expression subgroup. G-CIMP-positive samples are indicated as red data points and G-CIMP-negative samples are indicated as black data points. Median age at diagnosis is indicated for each subgroup by a horizontal solid black line. D–F) Kaplan-Meier survival curves for GBM methylation and gene expression subtypes. In each plot, the percent probability of survival is plotted versus time since diagnosis in weeks. All samples with survival data greater than five years were censored. D) Kaplan-Meier survival curves among the four GBM expression subtypes. Proneural tumors are in blue, Neural tumors are in green, Classical tumors are in red, and Mesenchymal tumors are in gold. E) Kaplan-Meier survival curves between the three DNA methylation clusters. Cluster 1 tumors are in red, cluster 2 tumors are in blue and cluster 3 tumors are in green. F) Kaplan-Meier survival curves between proneural G-CIMP-positive, proneural G-CIMP-negative and all non-proneural GBM tumors. Proneural G-CIMP-positive tumors are in red, proneural G-CIMP-negative tumors are in blue and all non-proneural GBM tumors are in black. See also Figure S2.
Figure 3. Significant regions of copy number variation in the G-CIMP genome
Copy number variation for 23,748 loci (across 22 autosomes and plotted in genomic coordinates along the _x_-axis) was analyzed using 61 proneural TCGA GBM tumors. Homozygous deletion is indicated in dark blue, hemizygous deletion in light blue, neutral/no change in white, gain in light red and high-level amplification in dark red. A) Copy number variation between proneural G-CIMP positive and G-CIMP negative tumors. The Cochran Armitage test for trend, percent total amplification/deletion and raw copy number values are listed. The – log10(FDR-adjusted P value) between G-CIMP-positive and G-CIMP negative proneurals is plotted along the _y_-axis in the “adjusted P-value pos vs. neg” panel. In this panel, red vertical lines indicate significance. Gene regions in 8q23.1-q24.3 and 10p15.2-11.21 are identified by asterisks and are highlighted in panels B and C, respectively. See also Figure S4 and Table S3.
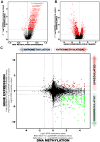
Figure 4. Comparison of transcriptome versus epigenetic differences between proneural G-CIMP-positive and G-CIMP-negative tumors
A) Volcano Plots of all CpG loci analyzed for G-CIMP association. The beta value difference in DNA Methylation between the proneural G-CIMP-positive and proneural G-CIMP-negative tumors is plotted on the x-axis, and the p-value for a FDR-corrected Wilcoxon signed-rank test of differences between the proneural G-CIMP-positive and proneural G-CIMP-negative tumors (−1* log10 scale) is plotted on the y-axis. Probes that are significantly different between the two subtypes are colored in red. B) Volcano plot for all genes analyzed on the Agilent gene expression platform. C) Starburst plot for comparison of TCGA Infinium DNA methylation and Agilent gene expression data normalized by copy number information for 11,984 unique genes. Log10(FDR-adjusted P value) is plotted for DNA methylation (_x-_axis) and gene expression (_y-_axis) for each gene. If a mean DNA methylation β-value or mean gene expression value is higher (greater than zero) in G-CIMP-positive tumors, −1 is multiplied to log10(FDR-adjusted P value), providing positive values. The dashed black lines indicates FDR-adjusted P value at 0.05. Data points in red indicate those that are significantly up- and down-regulated in their gene expression levels and significantly hypo- or hypermethylated in proneural G-CIMP-positive tumors. Data points in green indicate genes that are significantly down-regulated in their gene expression levels and hypermethylated in proneural G-CIMP-positive tumors compared to proneural G-CIMP-negative tumors. See also Figure S5 and Table S3.
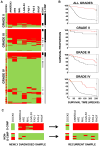
Figure 5. G-CIMP prevalence in grade II, III and IV gliomas using MethyLight
A) Methylation profiling of gliomas shows an association of CIMP status with tumor grade. Eight markers were tested for G-CIMP DNA methylation in 360 tumor samples. Each marker was coded as red if methylated and green if unmethylated. One of these markers (DOCK5) is unmethylated in CIMP, while the remaining seven markers show G-CIMP-specific hypermethylation. G-CIMP-positive status was determined if ≥6 of the 8 genes had G-CIMP-defining hyper- or hypomethylation. G-CIMP-positive status is indicated with a black line (right side of panel), and a grey line indicates non G-CIMP. Samples with an identified IDH1 mutation is indicated as a black line and samples with no known IDH1 mutation as a grey line. White line indicates unknown IDH1 status. B) Association of G-CIMP status with patient outcome stratified by tumor grade. G-CIMP-positive cases are indicated by red lines and the G-CIMP-negative cases are indicated by black lines in each Kaplan-Meier survival curve. C) Stability of G-CIMP over time in glioma patients. Fifteen samples from newly diagnosed tumors were tested for G-CIMP positivity using the eight-marker MethyLight panel. Eight tumors were classified as G-CIMP-positive (upper left panel), and seven tumors were classified as G-CIMP-negative (non-G-CIMP, lower left panel). Samples from a second procedure, ranging from 2–9 years after the initial resection, were also evaluated for the G-CIMP-positive cases (upper right panel), as well as for the non-G-CIMP cases (lower right panel). Each marker was coded as red if methylated and green if unmethylated.
Comment in
- To infinium, and beyond!
Wright KD, Gilbertson RJ. Wright KD, et al. Cancer Cell. 2010 May 18;17(5):419-20. doi: 10.1016/j.ccr.2010.04.020. Cancer Cell. 2010. PMID: 20478523
Similar articles
- CRISPR Editing of Mutant IDH1 R132H Induces a CpG Methylation-Low State in Patient-Derived Glioma Models of G-CIMP.
Moure CJ, Diplas BH, Chen LH, Yang R, Pirozzi CJ, Wang Z, Spasojevic I, Waitkus MS, He Y, Yan H. Moure CJ, et al. Mol Cancer Res. 2019 Oct;17(10):2042-2050. doi: 10.1158/1541-7786.MCR-19-0309. Epub 2019 Jul 10. Mol Cancer Res. 2019. PMID: 31292202 Free PMC article. - IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype.
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. Turcan S, et al. Nature. 2012 Feb 15;483(7390):479-83. doi: 10.1038/nature10866. Nature. 2012. PMID: 22343889 Free PMC article. - Molecular subtypes of glioma identified by genome-wide methylation profiling.
Kloosterhof NK, de Rooi JJ, Kros M, Eilers PH, Sillevis Smitt PA, van den Bent MJ, French PJ. Kloosterhof NK, et al. Genes Chromosomes Cancer. 2013 Jul;52(7):665-74. doi: 10.1002/gcc.22062. Epub 2013 Apr 30. Genes Chromosomes Cancer. 2013. PMID: 23629961 - Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma.
Agnihotri S, Aldape KD, Zadeh G. Agnihotri S, et al. Neurosurg Focus. 2014 Dec;37(6):E13. doi: 10.3171/2014.9.FOCUS14505. Neurosurg Focus. 2014. PMID: 25434382 Review. - The Cancer Genome Atlas expression profiles of low-grade gliomas.
Gonda DD, Cheung VJ, Muller KA, Goyal A, Carter BS, Chen CC. Gonda DD, et al. Neurosurg Focus. 2014 Apr;36(4):E23. doi: 10.3171/2012.12.focus12351. Neurosurg Focus. 2014. PMID: 24812719 Review.
Cited by
- Leveraging the replication-competent avian-like sarcoma virus/tumor virus receptor-A system for modeling human gliomas.
Kanvinde PP, Malla AP, Connolly NP, Szulzewsky F, Anastasiadis P, Ames HM, Kim AJ, Winkles JA, Holland EC, Woodworth GF. Kanvinde PP, et al. Glia. 2021 Sep;69(9):2059-2076. doi: 10.1002/glia.23984. Epub 2021 Feb 27. Glia. 2021. PMID: 33638562 Free PMC article. Review. - Methylation Profiling in Diffuse Gliomas: Diagnostic Value and Considerations.
Wenger A, Carén H. Wenger A, et al. Cancers (Basel). 2022 Nov 18;14(22):5679. doi: 10.3390/cancers14225679. Cancers (Basel). 2022. PMID: 36428772 Free PMC article. Review. - Up-regulation of microRNA-15b correlates with unfavorable prognosis and malignant progression of human glioma.
Pang C, Guan Y, Zhao K, Chen L, Bao Y, Cui R, Li G, Wang Y. Pang C, et al. Int J Clin Exp Pathol. 2015 May 1;8(5):4943-52. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26191187 Free PMC article. - Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients.
Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Qiu S, et al. J Transl Med. 2013 Jan 9;11:10. doi: 10.1186/1479-5876-11-10. J Transl Med. 2013. PMID: 23302469 Free PMC article. - Malignant glioma: lessons from genomics, mouse models, and stem cells.
Chen J, McKay RM, Parada LF. Chen J, et al. Cell. 2012 Mar 30;149(1):36-47. doi: 10.1016/j.cell.2012.03.009. Cell. 2012. PMID: 22464322 Free PMC article. Review.
References
- Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, Bigner DD. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. - PubMed
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. - PubMed
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. - PubMed
- Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 CA126561-03/CA/NCI NIH HHS/United States
- U24 CA143882/CA/NCI NIH HHS/United States
- U24 CA143882-01/CA/NCI NIH HHS/United States
- P50CA127001/CA/NCI NIH HHS/United States
- U24 CA126561/CA/NCI NIH HHS/United States
- P50 CA127001/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- U24 CA126561-03S1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous