Cohesinopathies, gene expression, and chromatin organization - PubMed (original) (raw)
Review
Cohesinopathies, gene expression, and chromatin organization
Tania Bose et al. J Cell Biol. 2010.
Abstract
The cohesin protein complex is best known for its role in sister chromatid cohesion, which is crucial for accurate chromosome segregation. Mutations in cohesin proteins or their regulators have been associated with human diseases (termed cohesinopathies). The developmental defects observed in these diseases indicate a role for cohesin in gene regulation distinct from its role in chromosome segregation. In mammalian cells, cohesin stably interacts with specific chromosomal sites and colocalizes with CTCF, a protein that promotes long-range DNA interactions, implying a role for cohesin in genome organization. Moreover, cohesin defects compromise the subnuclear position of chromatin. Therefore, defects in the cohesin network that alter gene expression and genome organization may underlie cohesinopathies.
Figures
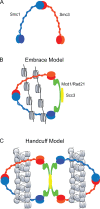
Figure 1.
The four subunits of the cohesin complex form a ring structure. The manner in which this ring interacts with DNA is a matter of debate. (A) Both Smc1 and Smc3 fold back on themselves to form intramolecular interactions. They associate to form a heterodimer via their hinge domains. (B) The Smc1-Smc3 heterodimer binds to Rad21 and Scc3, forming a ring. The cohesin ring is shown embracing two sister chromatids, drawn as 10-nm fibers, to mediate cohesion. (C) The cohesin ring may encircle a single sister chromatid and interact with a second ring containing the other sister to mediate cohesion. In this model, the DNA could be accommodated as a 30-nm fiber. The interaction between rings could be mediated through Rad21 and Scc3, a model known as the handcuff model.
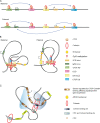
Figure 2.
The formation of chromatin loops is dependent on cohesin. (A) Schematic representation of long-distance interactions at the Igf2/H19 locus. Some interactions (blue arrows) occur on both alleles (biallelic), and some interactions are specific to the maternal or paternal allele. Specific CTCF cohesin–binding sites have been identified by 3C. Upstream of the IgF2 locus lies the differentially methylated region (DMR0), which includes a CTCF-binding site (CTCF AD). The centrally conserved DNase I hypersensitive site (CCD) lies between IgF2 and H19. A CTCF-binding region downstream of the H19 locus is denoted as CTCF DS. CTCF AD and CCD interactions occur on both alleles. (B) Looping may differ at the maternal and paternal Igf2/H19 locus. Cohesin and CTCF bind to the imprinting control region (ICR) when it is not methylated. CTCF AD and CCD interactions occur on both alleles. In the paternal allele, CTCF cohesin colocalization results in looping together the CTCF AD, CCD, and CTCF DS. Methylation of ICR causes CTCF DS to remain out of the loop, which in turn may cause activation of the IgF2 locus via interaction with the enhancer. The maternal allele carries an unmethylated copy of ICR that results in interaction between ICR and CTCF DS, which stops activation of IgF2 by the enhancer. (C) Schematic representation of the apolipoprotein locus. The AC2, AR1, and AC3 elements are bound to cohesin as measured by chromatin immunoprecipitation, but CTCF is only found associated with AC2 and AC3. 3C data indicate that the AC2, AR1, and AC3 elements interact with an enhancer in Hep3B cells, and the interaction is dependent on Rad21 and CTCF (Mishiro et al., 2009). The zone of influence of the enhancer element (yellow) is depicted in blue.
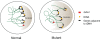
Figure 3.
Cohesin may contribute to subcellular localization of DNA sequences. tDNAs (yellow) cluster near the nucleolus (gray) in a budding yeast nucleus (Thompson et al., 2003). Genes (black) located adjacent to tDNAs can be silenced, and this depends on the proximity to the nucleolus (Wang et al., 2005). Recently, it was shown that strains bearing cohesinopathy mutations in either Eco1 (eco1-W216G) or Scc2 (scc2-D730V) lose tDNA clustering and tRNA gene–mediated silencing (Gard et al., 2009). GAL2 (red) is normally tethered to the nucleolus, but nucleolar morphology and GAL2 tethering is disrupted in the mutant backgrounds, and the induction of GAL2 is increased. Cohesin may contribute to tethering of tDNAs at a particular subcellular location, and this may affect the regulation of neighboring genes. The eco1-W216G mutation also causes defects in telomere clustering.
Similar articles
- The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review. - Cohesion and cohesin-dependent chromatin organization.
Nishiyama T. Nishiyama T. Curr Opin Cell Biol. 2019 Jun;58:8-14. doi: 10.1016/j.ceb.2018.11.006. Epub 2018 Dec 11. Curr Opin Cell Biol. 2019. PMID: 30544080 Review. - Cohesin ties up the genome.
Carretero M, Remeseiro S, Losada A. Carretero M, et al. Curr Opin Cell Biol. 2010 Dec;22(6):781-7. doi: 10.1016/j.ceb.2010.07.004. Epub 2010 Jul 31. Curr Opin Cell Biol. 2010. PMID: 20675112 Review. - Role of the DDX11 DNA Helicase in Warsaw Breakage Syndrome Etiology.
Santos D, Mahtab M, Boavida A, Pisani FM. Santos D, et al. Int J Mol Sci. 2021 Feb 25;22(5):2308. doi: 10.3390/ijms22052308. Int J Mol Sci. 2021. PMID: 33669056 Free PMC article. Review. - Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies.
Ball AR Jr, Chen YY, Yokomori K. Ball AR Jr, et al. Biochim Biophys Acta. 2014 Mar;1839(3):191-202. doi: 10.1016/j.bbagrm.2013.11.002. Epub 2013 Nov 22. Biochim Biophys Acta. 2014. PMID: 24269489 Free PMC article. Review.
Cited by
- Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions.
Biswas U, Wetzker C, Lange J, Christodoulou EG, Seifert M, Beyer A, Jessberger R. Biswas U, et al. PLoS Genet. 2013;9(12):e1003985. doi: 10.1371/journal.pgen.1003985. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385917 Free PMC article. - Cohesin recruits the Esco1 acetyltransferase genome wide to repress transcription and promote cohesion in somatic cells.
Rahman S, Jones MJ, Jallepalli PV. Rahman S, et al. Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11270-5. doi: 10.1073/pnas.1505323112. Epub 2015 Aug 24. Proc Natl Acad Sci U S A. 2015. PMID: 26305936 Free PMC article. - Cohesin SA2 is a sequence-independent DNA-binding protein that recognizes DNA replication and repair intermediates.
Countryman P, Fan Y, Gorthi A, Pan H, Strickland E, Kaur P, Wang X, Lin J, Lei X, White C, You C, Wirth N, Tessmer I, Piehler J, Riehn R, Bishop AJR, Tao YJ, Wang H. Countryman P, et al. J Biol Chem. 2018 Jan 19;293(3):1054-1069. doi: 10.1074/jbc.M117.806406. Epub 2017 Nov 24. J Biol Chem. 2018. PMID: 29175904 Free PMC article. - Chromosome domain architecture and dynamic organization of the fission yeast genome.
Mizuguchi T, Barrowman J, Grewal SI. Mizuguchi T, et al. FEBS Lett. 2015 Oct 7;589(20 Pt A):2975-86. doi: 10.1016/j.febslet.2015.06.008. Epub 2015 Jun 19. FEBS Lett. 2015. PMID: 26096785 Free PMC article. Review. - Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency.
Chen HS, Wikramasinghe P, Showe L, Lieberman PM. Chen HS, et al. J Virol. 2012 Sep;86(17):9454-64. doi: 10.1128/JVI.00787-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740398 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources