Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation - PubMed (original) (raw)
Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation
Yijian Rao et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Members of the Bin/amphiphysin/Rvs (BAR) domain protein superfamily are involved in membrane remodeling in various cellular pathways ranging from endocytic vesicle and T-tubule formation to cell migration and neuromorphogenesis. Membrane curvature induction and stabilization are encoded within the BAR or Fer-CIP4 homology-BAR (F-BAR) domains, alpha-helical coiled coils that dimerize into membrane-binding modules. BAR/F-BAR domain proteins often contain an SH3 domain, which recruits binding partners such as the oligomeric membrane-fissioning GTPase dynamin. How precisely BAR/F-BAR domain-mediated membrane deformation is regulated at the cellular level is unknown. Here we present the crystal structures of full-length syndapin 1 and its F-BAR domain. Our data show that syndapin 1 F-BAR-mediated membrane deformation is subject to autoinhibition by its SH3 domain. Release from the clamped conformation is driven by association of syndapin 1 SH3 with the proline-rich domain of dynamin 1, thereby unlocking its potent membrane-bending activity. We hypothesize that this mechanism might be commonly used to regulate BAR/F-BAR domain-induced membrane deformation and to potentially couple this process to dynamin-mediated fission. Our data thus suggest a structure-based model for SH3-mediated regulation of BAR/F-BAR domain function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
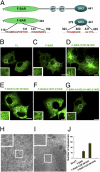
Syndapin 1 F-BAR but not full-length syndapin 1 forms membrane tubules in living cells. (A) Schematic representation of the domain organization of syndapin 1 and of the various truncations and mutants used in this study. (B_–_G) Live-cell spinning-disk confocal imaging of Cos7 cells expressing full-length (FL) eGFP-syndapin 1 wild-type or mutants. (B) Expression of full-length eGFP-syndapin 1 did not cause membrane tubulation. (C_–_G) Membrane tubules induced by expression of eGFP-syndapin 1 F-BAR wild-type (C) or (I122F/M123F) (D). Mutational inactivation of the amphipathic wedge loop I122T/M123Q (E) or the basic belt (K127E/K130E) (F) and (K145E/K146E/K148E) (G) eliminated the tubulating activity. Representative images from at least three independent experiments are shown. (Scale bars, 10 μm.) (H and I) Representative electron micrographs of Cos7 cells expressing full-length eGFP-syndapin 1 (H) or eGFP-syndapin 1 F-BAR (I). Note the presence of numerous F-BAR-induced membrane tubules in I. (Scale bar, 0.5 μm.) (J) Quantification of the number of eGFP-positive tubules in Cos7 cells expressing syndapin 1 variants. Given are the mean number of tubules (±SE) per cell (n = 3 for each syndapin 1 variant) averaged for at least 20 cells.
Fig. 2.
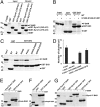
Structure of full-length syndapin 1. (A) Secondary-structure representation of syndapin 1. Protein coloration: gold, chain A; purple, chain B; green, chain D (the SH3 domain). The SH3 domain may be associated with either chain A or chain B of the F-BAR domain, but the lacking electron density for the tether disallows a definite chain assignment. A second SH3 domain found in the asymmetric unit which interacts with the F-BAR domain in an irrelevant manner is not shown in the model. N and C termini, the secondary-structure assignment, and the wedge loop protruding from the long helical region are labeled only on chain A. (B) Close-up view of the interface of the SH3 and F-BAR domains. β-Strands of the SH3 domain are labeled. Residues involved in the interaction are shown as a stick model. Polar contacts are presented as red dashed lines. (C) A proline-rich peptide (blue) is modeled by hand to the putative PxxP-binding groove in the syndapin 1 SH3 domain, corresponding to the peptide position in the SH3-peptide complex structures PDB ID codes 1W70 and 2DRK. (D) The protein surface is colored according to the electrostatic potential: red (negative) through white (neutral) to blue (positive). The lower panel shows an open-up view of the surface charge distribution between F-BAR and SH3. Residues mutated in functional studies are labeled.
Fig. 3.
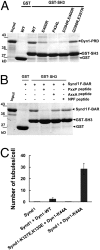
Complex formation between syndapin 1 F-BAR and SH3 regulates syndapin 1-induced membrane tubulation. (A) GST-syndapin 1 SH3 (379–441) or GST-syndapin 1 ΔF-BAR (340–441) bind to their F-BAR domain in vitro, whereas GST does not. (B) Mutation of a basic patch on the F-BAR domain (K145E/K146E/K148E) eliminates binding to SH3. (C) Mutations within the SH3 domain (D394R/E400R) and (Q396R/E397R) selectively impair binding to the F-BAR domain. By contrast, mutation of the proline-rich motif-binding site on SH3 (P434L) does not affect F-BAR association. Binding assays were done as described in Materials and Methods. Samples were analyzed by SDS/PAGE and staining with Coomassie blue. Input, 15% (A and B) or 10% (C) of the total amount of purified F-BAR used for the binding assay. (D) Cotransfection of wild-type (WT) but not mutant SH3 inhibits eGFP-syndapin 1 F-BAR-induced membrane tubulation. Shown are the mean number of tubules (±SE) per cell (n = 3 for each syndapin 1 variant) averaged for at least 20 cells. (E_–_G) In vitro binding assays. GST or GST-SH3 fusion proteins were immobilized on glutathione beads and incubated with purified His6-tagged endophilin 1 BAR (E), amphiphysin 1 BAR (F), or syndapin 1 F-BAR (G) domains. Input, 5% (E and F) or 3.5% (G) of the total amount of purified BAR domains used for the assay. Samples were analyzed by SDS/PAGE and staining with Coomassie blue.
Fig. 4.
Association of syndapin 1 SH3 with dynamin 1 releases its tubulating activity from autoinhibition. (A) The proline-rich domain of dynamin 1 (Dyn I-PRD) and syndapin 1 F-BAR show partially overlapping binding sites on syndapin 1 SH3. GST pull-down assays were done as described in Materials and Methods. (B) A dynamin 1-derived syndapin 1-binding wild-type (PxxP) but not an inactive mutant (AxxA) peptide (each at 100 μM) competes with F-BAR for binding to GST-SH3. A peptide derived from the syndapin 1 linker region (NPF) was taken as a further negative control. Samples were analyzed by SDS/PAGE and staining with Coomassie blue. Input, 15% (A and B) of the total amount of purified F-BAR domain used in the assay. (C) Expression of a GTPase-defective dynamin 1 mutant (K44A) unlocks a potent membrane-tubulating activity of eGFP-syndapin 1. Quantification of eGFP-syndapin 1-induced membrane tubules in Cos7 cells coexpressing either mRFP or mRFP-tagged dynamin 1 variants (wild-type or GTPase-defective mutant K44A). Shown are the mean number of tubules (±SE) per cell (n = 3 for each syndapin 1 variant) averaged for at least 20 cells.
Fig. 5.
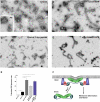
Syndapin 1-induced membrane tubulation reconstituted on liposomes in vitro. (A_–_D) Negative-stain electron microscopy. (A) Transmission electron microscope image of liposomes incubated with full-length (FL) syndapin 1. Inset shows liposomes without the protein. (B_–_D) Representative micrographs of liposomes incubated with syndapin 1 F-BAR (B), FL syndapin 1 with 170 μM dynamin peptide (C), and FL syndapin 1 (Q396R, E397R) (D). (Scale bar, 500 nm.) (E) Quantification of liposome tubulation in the experiments illustrated above. Note the significant increase in tubulation efficiency (P < 0.001, Student's t test) of FL syndapin 1 after addition of the dynamin peptide or upon mutation of its SH3 domain. (F) Hypothetical model for the cooperative role of BAR-SH3 domain-containing proteins and dynamin 1 in membrane deformation and fission. See text for details.
Similar articles
- Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2.
Neumann S, Schmid SL. Neumann S, et al. J Biol Chem. 2013 Aug 30;288(35):25119-25128. doi: 10.1074/jbc.M113.490474. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861397 Free PMC article. - Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Wang Q, et al. Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12700-5. doi: 10.1073/pnas.0902974106. Epub 2009 Jun 19. Proc Natl Acad Sci U S A. 2009. PMID: 19549836 Free PMC article. - Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis.
Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Tsujita K, et al. J Cell Biol. 2006 Jan 16;172(2):269-79. doi: 10.1083/jcb.200508091. J Cell Biol. 2006. PMID: 16418535 Free PMC article. - F-BAR family proteins, emerging regulators for cell membrane dynamic changes-from structure to human diseases.
Liu S, Xiong X, Zhao X, Yang X, Wang H. Liu S, et al. J Hematol Oncol. 2015 May 9;8:47. doi: 10.1186/s13045-015-0144-2. J Hematol Oncol. 2015. PMID: 25956236 Free PMC article. Review. - Syndapin--a membrane remodelling and endocytic F-BAR protein.
Quan A, Robinson PJ. Quan A, et al. FEBS J. 2013 Nov;280(21):5198-212. doi: 10.1111/febs.12343. Epub 2013 Jul 5. FEBS J. 2013. PMID: 23668323 Review.
Cited by
- Dynamin, a membrane-remodelling GTPase.
Ferguson SM, De Camilli P. Ferguson SM, et al. Nat Rev Mol Cell Biol. 2012 Jan 11;13(2):75-88. doi: 10.1038/nrm3266. Nat Rev Mol Cell Biol. 2012. PMID: 22233676 Free PMC article. Review. - Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis.
Giridharan SS, Cai B, Vitale N, Naslavsky N, Caplan S. Giridharan SS, et al. Mol Biol Cell. 2013 Jun;24(11):1776-90, S1-15. doi: 10.1091/mbc.E13-01-0026. Epub 2013 Apr 17. Mol Biol Cell. 2013. PMID: 23596323 Free PMC article. - Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst.
Teodoro RO, Pekkurnaz G, Nasser A, Higashi-Kovtun ME, Balakireva M, McLachlan IG, Camonis J, Schwarz TL. Teodoro RO, et al. EMBO J. 2013 Jul 17;32(14):2039-55. doi: 10.1038/emboj.2013.147. Epub 2013 Jun 28. EMBO J. 2013. PMID: 23812009 Free PMC article. - Membrane curvature and its generation by BAR proteins.
Mim C, Unger VM. Mim C, et al. Trends Biochem Sci. 2012 Dec;37(12):526-33. doi: 10.1016/j.tibs.2012.09.001. Epub 2012 Oct 8. Trends Biochem Sci. 2012. PMID: 23058040 Free PMC article. Review. - Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2.
Goretzki B, Glogowski NA, Diehl E, Duchardt-Ferner E, Hacker C, Gaudet R, Hellmich UA. Goretzki B, et al. Structure. 2018 Dec 4;26(12):1583-1593.e5. doi: 10.1016/j.str.2018.08.002. Epub 2018 Sep 20. Structure. 2018. PMID: 30244966 Free PMC article.
References
- Hatzakis NS, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5:835–841. - PubMed
- Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases