Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism - PubMed (original) (raw)
Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism
Leah K Hutnick et al. J Biol Chem. 2010.
Abstract
In defense of deleterious retrotransposition of intracisternal A particle (IAP) elements, IAP loci are heavily methylated and silenced in mouse somatic cells. To determine whether IAP is also repressed in pluripotent stem cells by DNA methylation, we examined IAP expression in demethylated mouse embryonic stem cells (mESCs) and epiblast-derived stem cells. Surprisingly, in demethylated ESC cultures carrying mutations of DNA methyltransferase I (Dnmt1), no IAP transcripts and proteins are detectable in undifferentiated Oct4(+) ESCs. In contrast, approximately 3.6% of IAP-positive cells are detected in Oct4(-) Dnmt1(-/-) cells, suggesting that the previously observed increase in IAP transcripts in the population of Dnmt1(-/-) ESCs could be accounted for by this subset of Oct4(-) Dnmt1(-/-) ESCs undergoing spontaneous differentiation. Consistent with this possibility, a dramatic increase of IAP mRNA (>100-fold) and protein expression was observed in Dnmt1(-/-) ESC cultures upon induction of differentiation through the withdrawal of leukemia-inhibitory factor for 6 or more days. Interestingly, both mRNAs and proteins of IAP can be readily detected in demethylated Oct4(+) epiblast-derived stem cells as well as differentiated mouse embryo fibroblasts, neurons, and glia upon conditional Dnmt1 gene deletion. These data suggest that mESCs are a unique stem cell type possessing a DNA methylation-independent IAP repression mechanism. This methylation-independent mechanism does not involve Dicer-mediated action of microRNAs or RNA interference because IAP expression remains repressed in Dnmt1(-/-); Dicer(-/-) double mutant ESCs. We suggest that mESCs possess a unique DNA methylation-independent mechanism to silence retrotransposons to safeguard genome stability while undergoing rapid cell proliferation for self-renewal.
Figures
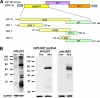
FIGURE 1.
IAP protein is only detected in lysates of E18.5 _Dnmt1_−/− mutant brain. A, depiction of protein structural domains of the full-length IAP clone MIA14. A deletion series of the gag peptide domain, which most closely corresponds to the p73 peptide used to generate the original IAP antibody (45), was cloned to create GST fusion peptides. B, immunoblotting revealing IAP expression in control (Con) and E18.5 _Nestin_-Cre; Dnmt1 conditional mutant brain lysate (Mut) of the original p73 serum before purification (anti-p73, left blot) and after column purification using the IAP2-GST fusion peptide (anti-p73, middle blot). Our IAP2 polyclonal antibody also strongly recognizes the 73-kDa gag protein (anti-IAP2, right blot) from mutant brain lysate (32). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
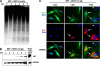
FIGURE 2.
IAP protein reactivation is tightly linked to the onset of DNA demethylation in MEFs. A, Dnmt1 2lox/2lox MEFs were infected with MSCV-Cre, and genomic demethylation was monitored using restriction enzyme digestion with methyl-sensitive HpaII followed be Southern blot analysis of IAP repetitive elements. Genomic demethylation appears 4 days after infection (DIV). B, immunoblotting shows that IAP protein expression appears 5 days postinfection with MSCV-Cre; thus, the IAP protein expression occurs after significant genome demethylation. The far right lane is a positive control for IAP proteins detected in the brain of E18.5 _Nestin_-Cre; Dnmt1 conditional knockouts (Brain E18.5 MUT) (29). C, IAP immunochemistry (red) colocalizes with MSCV-Cre-GFP-infected cells (green) once the genome becomes significantly demethylated after DNMT1 deletion. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; DAPI, 4′,6-diamidino-2-phenylindole.
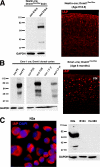
FIGURE 3.
Detection of IAP protein expression in demethylated somatic cells. A, left, immunoblotting using anti-IAP2 shows IAP protein in control and E18.5 _Nestin_-Cre; Dnmt1 conditional mutant brain lysates. Right, IAP immunoreactivity in E18.5 dorsal cortex of E18.5 _Nestin_-Cre; Dnmt1 mutant mice (29). B, left, Western blot analysis of IAP protein expression in control and _Emx1_-Cre; Dnmt1 cortical lysate from neonate P0 to 1.5 years old. Right, IAP immunoreactivity in the dorsal cortex (ctx) of 6-month-old _Emx1_-Cre; Dnmt1 mutant mice (9). C, immunocytochemistry for IAP protein in cultured N2a neuroblastoma cell lines. Western blot showing IAP expression in N2a cells but not in other types of neuroblastoma cells, including B104 and Hs 683 from ATCC. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; DAPI, 4′,6-diamidino-2-phenylindole.
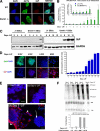
FIGURE 4.
Expression of IAP protein is suppressed in _Dnmt1_−/− mESCs and is only reactivated upon partial differentiation coupled with dramatic increases in IAP mRNAs. A, IAP protein is not expressed in undifferentiated _Dnmt1_−/− mESCs, although _Dnmt1_−/− cells possess less than 22% of normal genomic DNA methylation levels (28). Scale bar, 25 μm. B, upon induction of differentiation of _Dnmt1_−/− (c/c) mESCs in the absence of LIF treatment, IAP mRNA levels increase dramatically over the time course of 6–9 days in culture. No induction of IAP expression was observed in wild type control (J1) mESCs. C, IAP protein levels in the heterogeneous lysate of _Dnmt1_−/− cells were also detected after 6 days of LIF withdrawal, reaching a detectable threshold by Western blot. The blot on the left is a longer exposure to show detectable IAP expression at Day 6 of LIF withdrawal. D, IAP immunocytochemistry shows that only Oct4-negative cells express IAP protein in _Dnmt1_−/− ESCs. IAP-expressing cells were detected in a small population (under 10%) at Day 0–4 of differentiation but reached peak levels at Day 6–9 upon the induction of differentiation by LIF withdrawal. Scale bar, 50 μm. E, IAP FISH analysis reveals that both wild type (J1) and _Dnmt1_−/− (c/c) undifferentiated colonies at Day 0 of LIF withdrawal do not express IAP mRNA. Upon LIF withdrawal to induce differentiation, partially differentiated _Dnmt1_−/− cells migrating away from the colony express IAP RNA (arrowheads). _Dnmt1_−/− cells with undifferentiated colony morphology (arrow) do not express IAP mRNA. Scale bar, 25 μm. F, Northern blot analysis of the IAP gag coding region reveals the dramatic induction of IAP mRNA as LIF is withdrawn from culture. -Fold induction levels are assessed after normalization of 28 S rRNA bands from wild type (J1) and _Dnmt1_−/− (c/c) during the time course of differentiation. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; DAPI, 4′,6-diamidino-2-phenylindole. Error bars, S.E.
FIGURE 5.
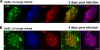
Hypomethylated EpiSCs rapidly induce IAP protein reactivation. A, photomicrographs of a representative colony 2 days after lentiviral cre recombinase delivery to Dnmt12lox/2lox EpiSCs. IAP immunoreactivity is present in Oct4-positive (blue) infected (GFP-positive, green) EpiSCs. Scale bar, 25 μm. B, 4 days after lentiviral cre recombinase delivery, colonies arising from infected EpiSCs show a homogenous population of Oct4/IAP/GFP-positive cells. Scale bar, 25 μm.
FIGURE 6.
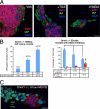
Inhibition of histone deacetylases and proteosome activities does not lead to IAP protein expression in undifferentiated _Dnmt1_−/− mESCs. A, HDAC inhibitor treatments of _Dnmt1_−/− mESCs in the presence of LIF were performed to question whether blocking histone deacetylation, thus promoting the active chromatin conformation, in _Dnmt1_−/− mESCs would lead to IAP protein expression in undifferentiated cells. Confocal images show that HDAC inhibitors do not reactivate IAP protein expression in _Dnmt1_−/− mESCs, suggesting that an alternative repressive mechanism is involved. Scale bar, 25 μm. B, cell counts show that HDAC inhibitors increase the number of IAP-positive colonies, which correlates with the known differentiating effect of HDAC inhibitors in cell culture. C, after 8 h of treatment with the proteosome inhibitor MG132, IAP protein was not visualized in Oct3/4-expressing _Dnmt1_−/− mESCs (confocal photomicrograph; scale bar, 25 μm). Error bars, S.E. DAPI, 4′,6-diamidino-2-phenylindole.
FIGURE 7.
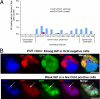
Clonal analysis of IAP expression in _Dicer_−/−; _Dnmt1_−/− mESCs. A, IAP protein reactivation was counted in individual clones after genotyping was performed. The vast majority of IAP-positive cells were Oct3/4-negative, thus indicating that only Dicer/Dnmt1 double mutant mESCs express IAP after spontaneous differentiation. Of note, two clones had a few cells expressing weak levels of IAP protein in Oct3/4-positive cells. The numbers on the x axis denote the clone number for the 3–6 individual clones with one of the following genotypes: Dnmt12lox; Dicer2lox (control); Dnmt11lox; Dicer2lox (_Dnmt1_−/− mutant cells); Dnmt11lox; Dicer1lox (_Dnmt1_−/−; _Dicer_−/− double mutant ESCs)). B, photomicrographs of clone 127, where differential levels of IAP are expressed. Strong IAP immunostaining is seen in differentiating (Oct3/4-negative) cells, whereas a weak perinuclear stain can be occasionally seen in few Oct3/4+ cells (arrow, bottom). Scale bar, 25 μm.
Similar articles
- DNMT1 can induce primary germ layer differentiation through de novo DNA methylation.
Ito T, Kubiura-Ichimaru M, Miura F, Tajima S, Surani MA, Ito T, Yamaguchi S, Tada M. Ito T, et al. Genes Cells. 2024 Jul;29(7):549-566. doi: 10.1111/gtc.13130. Epub 2024 May 29. Genes Cells. 2024. PMID: 38811355 Free PMC article. - Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing.
Gaudet F, Rideout WM 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Gaudet F, et al. Mol Cell Biol. 2004 Feb;24(4):1640-8. doi: 10.1128/MCB.24.4.1640-1648.2004. Mol Cell Biol. 2004. PMID: 14749379 Free PMC article. - DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells.
Ito T, Kubiura-Ichimaru M, Murakami Y, Bogutz AB, Lefebvre L, Suetake I, Tajima S, Tada M. Ito T, et al. PLoS One. 2022 Jan 5;17(1):e0262277. doi: 10.1371/journal.pone.0262277. eCollection 2022. PLoS One. 2022. PMID: 34986190 Free PMC article. - DNA methylation in embryonic stem cells.
Altun G, Loring JF, Laurent LC. Altun G, et al. J Cell Biochem. 2010 Jan 1;109(1):1-6. doi: 10.1002/jcb.22374. J Cell Biochem. 2010. PMID: 19899110 Free PMC article. Review. - Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?
Gunaratne PH. Gunaratne PH. Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
Cited by
- Drug Response-Related DNA Methylation Changes in Schizophrenia, Bipolar Disorder, and Major Depressive Disorder.
Zhou J, Li M, Wang X, He Y, Xia Y, Sweeney JA, Kopp RF, Liu C, Chen C. Zhou J, et al. Front Neurosci. 2021 May 13;15:674273. doi: 10.3389/fnins.2021.674273. eCollection 2021. Front Neurosci. 2021. PMID: 34054421 Free PMC article. Review. - Wnt/β-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells.
Theka I, Sottile F, Cammisa M, Bonnin S, Sanchez-Delgado M, Di Vicino U, Neguembor MV, Arumugam K, Aulicino F, Monk D, Riccio A, Cosma MP. Theka I, et al. Sci Rep. 2019 Jan 30;9(1):948. doi: 10.1038/s41598-018-37442-5. Sci Rep. 2019. PMID: 30700782 Free PMC article. - Characterization of DNA methylation and promoter activity of long terminal repeat elements of feline endogenous retrovirus RDRS C2a.
Shimode S, Yamamoto T. Shimode S, et al. Virus Genes. 2022 Feb;58(1):70-74. doi: 10.1007/s11262-021-01878-1. Epub 2021 Nov 17. Virus Genes. 2022. PMID: 34787790 - Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells.
Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, Meehan RR, Adams IR. Reichmann J, et al. PLoS Comput Biol. 2012;8(4):e1002486. doi: 10.1371/journal.pcbi.1002486. Epub 2012 Apr 26. PLoS Comput Biol. 2012. PMID: 22570599 Free PMC article. - Heterochromatic histone modifications at transposons in Xenopus tropicalis embryos.
van Kruijsbergen I, Hontelez S, Elurbe DM, van Heeringen SJ, Huynen MA, Veenstra GJC. van Kruijsbergen I, et al. Dev Biol. 2017 Jun 15;426(2):460-471. doi: 10.1016/j.ydbio.2016.08.031. Epub 2016 Sep 14. Dev Biol. 2017. PMID: 27639284 Free PMC article.
References
- Bestor T. H. (2000) Hum. Mol. Genet. 9, 2395–2402 - PubMed
- Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 - PubMed
- Robertson K. D., Wolffe A. P. (2000) Nat. Rev. Genet. 1, 11–19 - PubMed
- Jaenisch R., Bird A. (2003) Nat. Genet. 33, 245–254 - PubMed
- Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials