Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group - PubMed (original) (raw)
. 2010 May 20;28(15):2529-37.
doi: 10.1200/JCO.2009.23.4732. Epub 2010 Apr 20.
Alexander Kohlmann, Lothar Wieczorek, Giuseppe Basso, Geertruy Te Kronnie, Marie-Christine Béné, John De Vos, Jesus M Hernández, Wolf-Karsten Hofmann, Ken I Mills, Amanda Gilkes, Sabina Chiaretti, Sheila A Shurtleff, Thomas J Kipps, Laura Z Rassenti, Allen E Yeoh, Peter R Papenhausen, Wei-Min Liu, P Mickey Williams, Robin Foà
Affiliations
- PMID: 20406941
- PMCID: PMC5569671
- DOI: 10.1200/JCO.2009.23.4732
Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group
Torsten Haferlach et al. J Clin Oncol. 2010.
Abstract
Purpose: The Microarray Innovations in Leukemia study assessed the clinical utility of gene expression profiling as a single test to subtype leukemias into conventional categories of myeloid and lymphoid malignancies.
Methods: The investigation was performed in 11 laboratories across three continents and included 3,334 patients. An exploratory retrospective stage I study was designed for biomarker discovery and generated whole-genome expression profiles from 2,143 patients with leukemias and myelodysplastic syndromes. The gene expression profiling-based diagnostic accuracy was further validated in a prospective second study stage of an independent cohort of 1,191 patients.
Results: On the basis of 2,096 samples, the stage I study achieved 92.2% classification accuracy for all 18 distinct classes investigated (median specificity of 99.7%). In a second cohort of 1,152 prospectively collected patients, a classification scheme reached 95.6% median sensitivity and 99.8% median specificity for 14 standard subtypes of acute leukemia (eight acute lymphoblastic leukemia and six acute myeloid leukemia classes, n = 693). In 29 (57%) of 51 discrepant cases, the microarray results had outperformed routine diagnostic methods.
Conclusion: Gene expression profiling is a robust technology for the diagnosis of hematologic malignancies with high accuracy. It may complement current diagnostic algorithms and could offer a reliable platform for patients who lack access to today's state-of-the-art diagnostic work-up. Our comprehensive gene expression data set will be submitted to the public domain to foster research focusing on the molecular understanding of leukemias.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures
Fig 1.
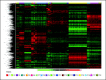
Supervised hierarchical clustering. The exploratory whole-genome clustering analysis was performed for all classes (C1 to C18 in ascending order) including 2,096 samples from stage I. For every class pair, the top 100 differentially expressed probes sets with the largest absolute values of t statistic were selected. The union of these sets contained 3,556 probe sets used in the clustering.
Fig 2.
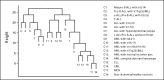
Exploratory margin tree analysis. Margin tree classification is a supervised multiclass support vector machine classification method. The margin tree program was applied to the stage I data set of 2,096 samples, characterized by their 18 class subtype labels (C1 to C18), and was based on 54,630 probe sets. B-ALL, B-cell acute lymphoblastic leukemia; MLL, myeloid/lymphoid or mixed-lineage leukemia; pre, precursor; c-ALL, childhood acute lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; kt., karyotype; abn., abnormality; CLL, chronic lymphocytic leukemia; CML, chronic myelogenous leukemia; MDS, myelodysplastic syndrome.
Fig 3.
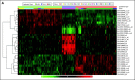
Virtual immunophenotypes for 1,292 acute leukemia specimens from stage I of the Microarray Innovations in Leukemia study. (A) Microarray gene expression signal intensities of 21 differentiation antigens currently tested in flow cytometry for the diagnosis of leukemia represented by 32 probe sets. (B) Gene expression intensities for CD3G, CD19, CD33, and HLA-DRA. Each dot represents the data from a single microarray profile. B-ALL, B-cell acute lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia; AML, acute myeloid leukemia.
Fig 3.
Virtual immunophenotypes for 1,292 acute leukemia specimens from stage I of the Microarray Innovations in Leukemia study. (A) Microarray gene expression signal intensities of 21 differentiation antigens currently tested in flow cytometry for the diagnosis of leukemia represented by 32 probe sets. (B) Gene expression intensities for CD3G, CD19, CD33, and HLA-DRA. Each dot represents the data from a single microarray profile. B-ALL, B-cell acute lymphoblastic leukemia; T-ALL, T-cell acute lymphoblastic leukemia; AML, acute myeloid leukemia.
Fig A1.
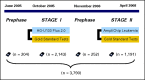
Overview of the Microarray Innovations in Leukemia (MILE) study timeline and corresponding number of microarray analyses for each study phase. There were two stages in the MILE research study: a retrospective biomarker discovery phase (stage I) using commercially available whole-genome microarrays (HG-U133 Plus 2.0, Affymetrix, Santa Clara, CA), and the independent validation phase (stage II) that was performed in a prospective manner using a focused and newly designed custom chip (AmpliChip Leukemia, Roche Molecular Systems, Pleasanton, CA). Before each stage of the study, designated laboratory operators were trained during a prephase on the corresponding sample preparation protocol and microarray workflow.
Fig A2.
Microarray analysis. Each center was provided with identical laboratory supplies and equipment for microarray analysis, including reagent kits, enzymes, spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE), and two heat block instruments (ThermoStat Plus, Eppendorf, Hamburg, Germany; Hybex incubation system, SciGene, Sunnyvale, CA). Each laboratory designated specific test operators, and all operators were individually trained during a 5-day course on a standardized sample preparation protocol for microarray analysis using commercially available total RNA from the MCF-7 and HepG2 cell lines (Ambion, Austin, TX). Detailed results on this prephase program have been published elsewhere.
Fig A3.
Stage II samples with discrepancies between gold-standard methods and array prediction. The figure summarizes 51 (7.4%) of 693 cases of acute leukemias from stage II where re-examination of the microarray classification result had led to a revision of the laboratories′ originally submitted gold standard. In 22 (43%) of 51 cases, errors were considered to be human errors (Fig A3A). These 43% of cases are split between 13 cases where sample information had been entered with erroneous information into the study database and nine cases where the sample karyotype category had to be reclassified after independent expert review following the definition from Schoch et al (Fig A3B). However, in a larger group of 29 (57%) of 51 cases, the microarray prediction turned out to be better than the gold standard. These 29 cases were further distributed between two subcategories, namely, (i) material was re-tested by RT-PCR or FISH assays (14 cases), and (ii) re-evaluation of morphology, DNA index, or other information from the initial diagnostic report (15 cases). Table A2 gives a more detailed overview on the type of tie-breaking molecular assay to resolve discrepancies between original gold standard (GS) and the microarray prediction result.
Fig A4.
Cases of AML misclassified as T-ALL. (A,C) Cross-validation training dataset from stage I. (B,D) Independent test data from stage II. There are recent data available that AML cases with CEBPA silencing have T-lineage features and exhibit a distinctive gene expression profile (Wouters BJ et al: Blood 113:3088-3091, 2009). Hypermethylation of the proximal CEBPA promoter was identified in a small subset of AMLs with low CEBPA mRNA expression. Here, an analysis was performed to investigate the gene expression intensity of CEBPA and a correlation with misclassification of AML cases by the microarray prediction algorithm into the category of T-ALL. The identity of the corresponding cases is given in Table A3.
Comment in
- Genetics: Gene-expression profiling in leukemia--a valuable diagnostic tool.
Richards L. Richards L. Nat Rev Clin Oncol. 2010 Aug;7(8):422. doi: 10.1038/nrclinonc.2010.112. Nat Rev Clin Oncol. 2010. PMID: 20700897 No abstract available.
Similar articles
- Gene expression profiling as a tool for the diagnosis of acute leukemias.
Haferlach T, Kohlmann A, Kern W, Hiddemann W, Schnittger S, Schoch C. Haferlach T, et al. Semin Hematol. 2003 Oct;40(4):281-95. doi: 10.1016/s0037-1963(03)00193-8. Semin Hematol. 2003. PMID: 14582079 Review. - Diagnosis and genetic subtypes of leukemia combining gene expression and flow cytometry.
Basso G, Case C, Dell'Orto MC. Basso G, et al. Blood Cells Mol Dis. 2007 Sep-Oct;39(2):164-8. doi: 10.1016/j.bcmd.2007.05.004. Epub 2007 Jun 27. Blood Cells Mol Dis. 2007. PMID: 17588788 - Cross-platform classification in microarray-based leukemia diagnostics.
Nilsson B, Andersson A, Johansson M, Fioretos T. Nilsson B, et al. Haematologica. 2006 Jun;91(6):821-4. Haematologica. 2006. PMID: 16769585 - Blood-based transcriptomics: leukemias and beyond.
Staratschek-Jox A, Classen S, Gaarz A, Debey-Pascher S, Schultze JL. Staratschek-Jox A, et al. Expert Rev Mol Diagn. 2009 Apr;9(3):271-80. doi: 10.1586/erm.09.9. Expert Rev Mol Diagn. 2009. PMID: 19379085 Review. - Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status.
Andersson A, Ritz C, Lindgren D, Edén P, Lassen C, Heldrup J, Olofsson T, Råde J, Fontes M, Porwit-Macdonald A, Behrendtz M, Höglund M, Johansson B, Fioretos T. Andersson A, et al. Leukemia. 2007 Jun;21(6):1198-203. doi: 10.1038/sj.leu.2404688. Epub 2007 Apr 5. Leukemia. 2007. PMID: 17410184
Cited by
- Transcriptome Analysis of Minimal Residual Disease in Subtypes of Pediatric B Cell Acute Lymphoblastic Leukemia.
Sitthi-Amorn J, Herrington B, Megason G, Pullen J, Gordon C, Hogan S, Koganti T, Hicks C. Sitthi-Amorn J, et al. Clin Med Insights Oncol. 2015 May 24;9:51-60. doi: 10.4137/CMO.S17049. eCollection 2015. Clin Med Insights Oncol. 2015. PMID: 26056509 Free PMC article. - Synergism between IL7R and CXCR4 drives BCR-ABL induced transformation in Philadelphia chromosome-positive acute lymphoblastic leukemia.
Abdelrasoul H, Vadakumchery A, Werner M, Lenk L, Khadour A, Young M, El Ayoubi O, Vogiatzi F, Krämer M, Schmid V, Chen Z, Yousafzai Y, Cario G, Schrappe M, Müschen M, Halsey C, Mulaw MA, Schewe DM, Hobeika E, Alsadeq A, Jumaa H. Abdelrasoul H, et al. Nat Commun. 2020 Jun 24;11(1):3194. doi: 10.1038/s41467-020-16927-w. Nat Commun. 2020. PMID: 32581241 Free PMC article. - Enhancer-activated RET confers protection against oxidative stress to KMT2A-rearranged acute myeloid leukemia.
Frett B, Stephens KE, Koss B, Melnyk S, Farrar J, Saha D, Roy Choudhury S. Frett B, et al. Cancer Sci. 2024 Mar;115(3):963-973. doi: 10.1111/cas.16069. Epub 2024 Jan 16. Cancer Sci. 2024. PMID: 38226414 Free PMC article. - The ferroptosis landscape in acute myeloid leukemia.
Ma Z, Ye W, Huang X, Li X, Li F, Lin X, Hu C, Wang J, Jin J, Zhu B, Huang J. Ma Z, et al. Aging (Albany NY). 2023 Nov 29;15(22):13486-13503. doi: 10.18632/aging.205257. Epub 2023 Nov 29. Aging (Albany NY). 2023. PMID: 38032290 Free PMC article. - BubR1 is frequently repressed in acute myeloid leukemia and its re-expression sensitizes cells to antimitotic therapy.
Schnerch D, Schmidts A, Follo M, Udi J, Felthaus J, Pfeifer D, Engelhardt M, Wäsch R. Schnerch D, et al. Haematologica. 2013 Dec;98(12):1886-95. doi: 10.3324/haematol.2013.087452. Epub 2013 Jun 28. Haematologica. 2013. PMID: 23812934 Free PMC article.
References
- Armstrong SA Staunton JE Silverman LB, etal: MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia Nat Genet 30:41–47,2002 - PubMed
- Chiaretti S Li X Gentleman R, etal: Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival Blood 103:2771–2778,2004 - PubMed
- Golub TR Slonim DK Tamayo P, etal: Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring Science 286:531–537,1999 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases