Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples - PubMed (original) (raw)
Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples
Christian L Lauber et al. FEMS Microbiol Lett. 2010 Jun.
Abstract
Storage conditions are considered to be a critical component of DNA-based microbial community analysis methods. However, whether differences in short-term sample storage conditions impact the assessment of bacterial community composition and diversity requires systematic and quantitative assessment. Therefore, we used barcoded pyrosequencing of bacterial 16S rRNA genes to survey communities, harvested from a variety of habitats [soil, human gut (feces) and human skin] and subsequently stored at 20, 4, -20 and -80 degrees C for 3 and 14 days. Our results indicate that the phylogenetic structure and diversity of communities in individual samples were not significantly influenced by the storage temperature or the duration of storage. Likewise, the relative abundances of most taxa were largely unaffected by temperature even after 14 days of storage. Our results indicate that environmental factors and biases in molecular techniques likely confer greater amounts of variation to microbial communities than do differences in short-term storage conditions, including storage for up to 2 weeks at room temperature. These results suggest that many samples collected and stored under field conditions without refrigeration may be useful for microbial community analyses.
Figures
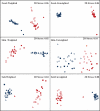
Fig.1
Non-metric Multidimensional Scaling (NMDS) plots of UniFrac weighted and unweighted pairwise distances. Overall community composition was not affected by temperature or duration of storage for weighted UniFrac distances (_P_> 0.1 in all cases). Length of storage significantly affected the skin communities for the unweighted UniFrac metric (P = 0.02). The remaining unweighted UniFrac distances were not significantly different by day or temperature. Blue=sample 1, red = sample 2. Open symbols = Day 3, closed symbols = Day 14. ▲= 20°C, ■ = 4°C, ● = −20°C, ◆ = −80°C.
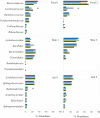
Fig. 2
The relative abundance of various bacterial taxa in fecal, skin and soil samples after 14 days of storage. Bars represent the mean abundance of each group at each temperature in descending order (e.g. 20, 4, −20, and −80°C) with 1 standard error of the mean. Abundances of bacterial taxa were classified to family for the fecal samples and to order for the skin and soil samples using the RDPII nomenclature. Differences in relative abundances due to storage temperature were assessed within individual samples using the Kruskall-Wallis test. Asterisk (*) indicates _P_- values < = 0.05.
Similar articles
- Storage conditions of intestinal microbiota matter in metagenomic analysis.
Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. Cardona S, et al. BMC Microbiol. 2012 Jul 30;12:158. doi: 10.1186/1471-2180-12-158. BMC Microbiol. 2012. PMID: 22846661 Free PMC article. - Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles.
Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, Hayden MK; CDC Prevention Epicenters Program. Bassis CM, et al. BMC Microbiol. 2017 Mar 31;17(1):78. doi: 10.1186/s12866-017-0983-9. BMC Microbiol. 2017. PMID: 28359329 Free PMC article. - Effects of Specimen Collection Methodologies and Storage Conditions on the Short-Term Stability of Oral Microbiome Taxonomy.
Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Luo T, et al. Appl Environ Microbiol. 2016 Aug 30;82(18):5519-29. doi: 10.1128/AEM.01132-16. Print 2016 Sep 15. Appl Environ Microbiol. 2016. PMID: 27371581 Free PMC article. - Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags.
Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, Li H, Bushman FD. Wu GD, et al. BMC Microbiol. 2010 Jul 30;10:206. doi: 10.1186/1471-2180-10-206. BMC Microbiol. 2010. PMID: 20673359 Free PMC article. - Time between collection and storage significantly influences bacterial sequence composition in sputum samples from cystic fibrosis respiratory infections.
Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hafiz T, Hoffman LR, Carroll MP, Parkhill J, Bruce KD, van der Gast CJ. Cuthbertson L, et al. J Clin Microbiol. 2014 Aug;52(8):3011-6. doi: 10.1128/JCM.00764-14. Epub 2014 Jun 11. J Clin Microbiol. 2014. PMID: 24920767 Free PMC article.
Cited by
- Characterization of the gut microbiome in epidemiologic studies: the multiethnic cohort experience.
Fu BC, Randolph TW, Lim U, Monroe KR, Cheng I, Wilkens LR, Le Marchand L, Hullar MA, Lampe JW. Fu BC, et al. Ann Epidemiol. 2016 May;26(5):373-9. doi: 10.1016/j.annepidem.2016.02.009. Epub 2016 Mar 8. Ann Epidemiol. 2016. PMID: 27039047 Free PMC article. Review. - Time of sample collection is critical for the replicability of microbiome analyses.
Allaband C, Lingaraju A, Flores Ramos S, Kumar T, Javaheri H, Tiu MD, Dantas Machado AC, Richter RA, Elijah E, Haddad GG, Leone VA, Dorrestein PC, Knight R, Zarrinpar A. Allaband C, et al. Nat Metab. 2024 Jul;6(7):1282-1293. doi: 10.1038/s42255-024-01064-1. Epub 2024 Jul 1. Nat Metab. 2024. PMID: 38951660 Free PMC article. - Comparison between 16S rRNA and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota.
Bars-Cortina D, Ramon E, Rius-Sansalvador B, Guinó E, Garcia-Serrano A, Mach N, Khannous-Lleiffe O, Saus E, Gabaldón T, Ibáñez-Sanz G, Rodríguez-Alonso L, Mata A, García-Rodríguez A, Obón-Santacana M, Moreno V. Bars-Cortina D, et al. BMC Genomics. 2024 Jul 29;25(1):730. doi: 10.1186/s12864-024-10621-7. BMC Genomics. 2024. PMID: 39075388 Free PMC article. - A jungle in there: bacteria in belly buttons are highly diverse, but predictable.
Hulcr J, Latimer AM, Henley JB, Rountree NR, Fierer N, Lucky A, Lowman MD, Dunn RR. Hulcr J, et al. PLoS One. 2012;7(11):e47712. doi: 10.1371/journal.pone.0047712. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23144827 Free PMC article. - Experimental and analytical tools for studying the human microbiome.
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Kuczynski J, et al. Nat Rev Genet. 2011 Dec 16;13(1):47-58. doi: 10.1038/nrg3129. Nat Rev Genet. 2011. PMID: 22179717 Free PMC article. Review.
References
- Clarke KR, Warwick RM. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Marine Ecology-Progress Series. 2001;216:265–278.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases