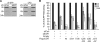E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair - PubMed (original) (raw)
E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair
Ruifeng Guo et al. J Biol Chem. 2010.
Abstract
The E2F1 transcription factor is a well known regulator of cell proliferation and apoptosis, but its role in the DNA damage response is less clear. Using a local UV irradiation technique and immunofluorescence staining, E2F1 is shown to accumulate at sites of DNA damage. Localization of E2F1 to UV-damaged DNA requires the ATM and Rad3-related (ATR) kinase and serine 31 of E2F1 but not an intact DNA binding domain. E2F1 deficiency does not appear to affect the expression of nucleotide excision repair (NER) factors, such as XPC and XPA. However, E2F1 depletion does impair the recruitment of NER factors to sites of damage and reduces the efficiency of DNA repair. E2F1 mutants unable to bind DNA or activate transcription retain the ability to stimulate NER. These findings demonstrate that E2F1 has a direct, non-transcriptional role in DNA repair involving increased recruitment of NER factors to sites of damage.
Figures
FIGURE 1.
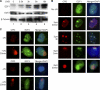
E2F1 accumulates at sites of UV-induced DNA damage. A, primary NHFs were irradiated with 500 J/m2 of UVB or mock treated (lane 1) and harvested at the indicated times post-irradiation. Western blot analysis was performed on whole cell extracts using antibodies specific for E2F1, E2F3, or β-tubulin. B, NHFs were untreated or locally irradiated with 50 or 100 J/m2 of UVC through polycarbonate filters with pores of 3 or 8 μm as indicated. 2 min, 30 min, or 3 h post-irradiation, cells were fixed and stained for CPD (red) and E2F1 (green) by indirect immunofluorescence. Cells were counterstained with DAPI to show nuclei and images were digitally recorded. C, NHFs were untreated or locally irradiated with 50 J/m2 of UVC and stained for CPD and E2F2 or E2F3 as described above.
FIGURE 2.
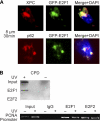
E2F1 co-localizes with NER factors and associates with damaged DNA. A, HeLa cells were transfected with plasmids expressing wild-type E2F1 fused to enhanced GFP. 24 h post-transfection, cells were exposed to 50 J/m2 of UVC through an 8-μm pore filter and stained 30 min later for XPC or p62 (red). Slides were counterstained with DAPI, and images were digitally recorded as previously described. B, NHFs were mock treated or exposed to 500 J/m2 of UVB and fixed with 1% formaldehyde for 15 min. Cells were lysed in the standard lysis buffer and chromatin immunoprecipitation was performed using antibody to either E2F1 or E2F2. The DNA in the precipitates was purified and analyzed by slot blotting for CPD photoproducts (top) or by PCR for the PCNA promoter region (bottom).
FIGURE 3.
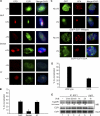
ATR and E2F1 serine 31 are required for the accumulation of E2F1 at sites of UV damage. A, primary NHFs, ATR-deficient human fibroblasts from a Seckel Syndrome patient (Seckel cells), and ATM-deficient human fibroblasts from an ataxia-telangiectasia patient (AT cells) were mock treated or exposed to 50 J/m2 of UVC through an 8-μm pore filter. 30 min post-irradiation, cells were fluorescently stained for CPD (red) and E2F1 (green) and counterstained with DAPI. B, 100 randomly selected cells were scored for co-localization of CPD and E2F1 in each of the cell types indicated. The average of three independent experiments is presented. *, indicates statistically significant difference from NHFs by Student's t test (p < 0.05). C, HeLa cells were transfected with plasmids expressing GFP fused to either wild-type E2F1 or E2F1 mutated at serine 31 (S31A). 24 h post-transfection, cells were exposed to 50 J/m2 of UVC through an 8-μm pore filter and 30 min later stained for XPA (red). Slides were then counterstained with DAPI, and images were digitally recorded as above. D, co-localization of XPA and GFP shown in C were scored and plotted as described above. The average of two independent experiments is presented. *, statistically significant difference (p < 0.05). E, HEK293 cells were untreated (lane 1) or exposed to 500 J/m2 of UVB and harvested at different time points post-exposure (lanes 2–5) as indicated. Co-immunoprecipitation was performed using antibody to E2F1 (Santa Cruz Biotechnology, KH95) (lanes 1–5) and normal mouse IgG as negative control (lane 6). The precipitate was subjected to Western blot analysis for TopBP1 (top panel) and E2F1 (middle panel). The input for E2F1 is shown by Western blot (lower panel). The -fold increase in TopBP1 association was calculated by determining the density of the TopBP1 band and normalizing to the intensity of the E2F1 input band.
FIGURE 4.
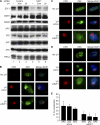
E2F1 deficiency impairs the recruitment of NER factors to sites of UV damage. A, NHFs were transfected with control siRNA (lanes 1–3) or siRNA specific to E2F1 (lanes 4–6). 24 h later, cells were mock treated (lanes 1 and 4) or exposed to 500 J/m2 of UVB and harvested at the indicated time points post-irradiation. Western blot analysis was performed using whole cell lysates and antibodies specific for E2F1 or the DNA repair proteins as indicated. B–D, NHFs were mock treated (upper panels) or transfected with control siRNA (siCon, middle panels) or with siRNA to E2F1 (lower panels). Transfected cells were locally irradiated with 50 J/m2 of UVC through a filter (middle and lower panels) and 30 min later fluorescently stained for CPD (red) and XPC, XPA, or p62 (green). Cells were counterstained with DAPI and digitally recorded as previously described. E, co-localization of XPC, XPA, and p62 with CPD shown in B–D were scored from three independent experiments. *, statistically significant difference between siCon- and siE2F1-transfected cells (p < 0.05).
FIGURE 5.
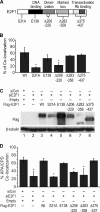
Mapping E2F1 domains required for E2F1 and XPA localization to sites of DNA damage. A, schematic diagram of E2F1 indicating sites of mutations. B, HCT116 cells were transiently transfected with either empty vector or the indicated versions of FLAG-tagged E2F1. Cells were then irradiated with 50 J/m2 of UVC through a filter and fluorescently stained for FLAG (green) and CPD (red) 15 min later. Cells were counterstained with DAPI. Co-localization of FLAG tag with CPD were scored as described. The average of two independent experiments is presented. *, statistically significant difference from wild type (p < 0.05). C, HCT116 cells were co-transfected with control siRNA (siCon, lane 1) or E2F1 siRNA specific to the 3′-untranslated region (lanes 2–8) and empty vector (lane 2) or plasmids expressing wild-type (lane 3) or different mutant versions of FLAG-tagged E2F1 as indicated. 24 h post transfection, Western blot analysis was performed for FLAG tag (top panel) and β-tubulin (bottom panel). D, HCT116 cells were transfected as in C. Following transfection, cells were exposed to 50 J/m2 of UVC through a filter and fluorescently stained for XPA (green) and CPD (red) 15 min post-irradiation. Cells were counterstained with DAPI. Rescue of XPA co-localization with CPD was scored as described. The average of two independent experiments is presented. *, statistically significant difference from cells co-transfected with siCon and empty vector (p < 0.05).
FIGURE 6.
Stimulation of GG-NER by E2F1 requires serine 31 but not functional DNA binding or transcriptional activation domains. A, NHFs were transfected with control siRNA or siRNA to E2F1 and 24 h later exposed to 500 J/m2 of UVB. Genomic DNA (0.5 μg) was taken immediately after UVB treatment (0 h) or 6 and 24 h post-irradiation and slot blotted. DNA damage was detected using antibodies specific for (6–4)PP or CPD. B, HCT116 cells were transfected as described in Fig. 5_C_. Cells were exposed to 500 J/m2 UVB and harvested immediately (0 h) or 6 and 24 h post-irradiation. Genomic DNA was extracted and immuno-slot-blot analysis was performed for (6–4)PP. The intensity of the bands was quantified by ImageJ software, and the results are plotted after normalization with total single strand DNA levels. The average from three independent experiments is presented. *, statistically significant difference from cells transfected with siE2F1 and rescued with empty vector (p < 0.05).
Similar articles
- The E2F1 transcription factor and RB tumor suppressor moonlight as DNA repair factors.
Manickavinayaham S, Velez-Cruz R, Biswas AK, Chen J, Guo R, Johnson DG. Manickavinayaham S, et al. Cell Cycle. 2020 Sep;19(18):2260-2269. doi: 10.1080/15384101.2020.1801190. Epub 2020 Aug 13. Cell Cycle. 2020. PMID: 32787501 Free PMC article. Review. - The role of the retinoblastoma/E2F1 tumor suppressor pathway in the lesion recognition step of nucleotide excision repair.
Lin PS, McPherson LA, Chen AY, Sage J, Ford JM. Lin PS, et al. DNA Repair (Amst). 2009 Jul 4;8(7):795-802. doi: 10.1016/j.dnarep.2009.03.003. Epub 2009 Apr 18. DNA Repair (Amst). 2009. PMID: 19376752 Free PMC article. - ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation.
Wu X, Shell SM, Liu Y, Zou Y. Wu X, et al. Oncogene. 2007 Feb 1;26(5):757-64. doi: 10.1038/sj.onc.1209828. Epub 2006 Jul 24. Oncogene. 2007. PMID: 16862173 Free PMC article. - Xeroderma Pigmentosa Group A (XPA), Nucleotide Excision Repair and Regulation by ATR in Response to Ultraviolet Irradiation.
Musich PR, Li Z, Zou Y. Musich PR, et al. Adv Exp Med Biol. 2017;996:41-54. doi: 10.1007/978-3-319-56017-5_4. Adv Exp Med Biol. 2017. PMID: 29124689 Free PMC article. Review.
Cited by
- The E2F1 transcription factor and RB tumor suppressor moonlight as DNA repair factors.
Manickavinayaham S, Velez-Cruz R, Biswas AK, Chen J, Guo R, Johnson DG. Manickavinayaham S, et al. Cell Cycle. 2020 Sep;19(18):2260-2269. doi: 10.1080/15384101.2020.1801190. Epub 2020 Aug 13. Cell Cycle. 2020. PMID: 32787501 Free PMC article. Review. - p53: Guardian of pancreatic epithelial identity.
Ghosh B, Leach SD. Ghosh B, et al. Cell Cycle. 2011 Jun 1;10(11):1717. doi: 10.4161/cc.10.11.15686. Epub 2011 Jun 1. Cell Cycle. 2011. PMID: 21597331 Free PMC article. No abstract available. - Metastasis is altered through multiple processes regulated by the E2F1 transcription factor.
Swiatnicki MR, Andrechek ER. Swiatnicki MR, et al. Sci Rep. 2021 May 4;11(1):9502. doi: 10.1038/s41598-021-88924-y. Sci Rep. 2021. PMID: 33947907 Free PMC article. - A new role for E2F1 in DNA repair: all for the greater good.
Degregori J. Degregori J. Cell Cycle. 2011 Jun 1;10(11):1716. doi: 10.4161/cc.10.11.15649. Epub 2011 Jun 1. Cell Cycle. 2011. PMID: 21532333 Free PMC article. No abstract available. - E2F1: A new role in the DNA damage response.
Cress WD. Cress WD. Cell Cycle. 2011 Jun 1;10(11):1718. doi: 10.4161/cc.10.11.15687. Epub 2011 Jun 1. Cell Cycle. 2011. PMID: 21543892 Free PMC article. No abstract available.
References
- Friedberg E. C. (2001) Nat. Rev. Cancer 1, 22–33 - PubMed
- Hanasoge S., Ljungman M. (2007) Carcinogenesis 28, 2298–2304 - PubMed
- Zou L., Elledge S. J. (2003) Science 300, 1542–1548 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 CA106603/CA/NCI NIH HHS/United States
- R01 CA079648/CA/NCI NIH HHS/United States
- K01-CA106603/CA/NCI NIH HHS/United States
- P30 ES007784/ES/NIEHS NIH HHS/United States
- ES007247/ES/NIEHS NIH HHS/United States
- T32 ES007247/ES/NIEHS NIH HHS/United States
- CA105345/CA/NCI NIH HHS/United States
- U01 CA105345/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- ES007784/ES/NIEHS NIH HHS/United States
- CA079648/CA/NCI NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous