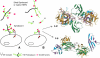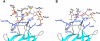Heparin-derived heparan sulfate mimics to modulate heparan sulfate-protein interaction in inflammation and cancer - PubMed (original) (raw)
Review
Heparin-derived heparan sulfate mimics to modulate heparan sulfate-protein interaction in inflammation and cancer
Benito Casu et al. Matrix Biol. 2010 Jul.
Abstract
The heparan sulfate (HS) chains of heparan sulfate proteoglycans (HSPG) are "ubiquitous" components of the cell surface and the extracellular matrix (EC) and play important roles in the physiopathology of developmental and homeostatic processes. Most biological properties of HS are mediated by interactions with "heparin-binding proteins" and can be modulated by exogenous heparin species (unmodified heparin, low molecular weight heparins, shorter heparin oligosaccharides and various non-anticoagulant derivatives of different sizes). Heparin species can promote or inhibit HS activities to different extents depending, among other factors, on how closely their structure mimics the biologically active HS sequences. Heparin shares structural similarities with HS, but is richer in "fully sulfated" sequences (S domains) that are usually the strongest binders to heparin/HS-binding proteins. On the other hand, HS is usually richer in less sulfated, N-acetylated sequences (NA domains). Some of the functions of HS chains, such as that of activating proteins by favoring their dimerization, often require short S sequences separated by rather long NA sequences. The biological activities of these species cannot be simulated by heparin, unless this polysaccharide is appropriately chemically/enzymatically modified or biotechnologically engineered. This mini review covers some information and concepts concerning the interactions of HS chains with heparin-binding proteins and some of the approaches for modulating HS interactions relevant to inflammation and cancer. This is approached through a few illustrative examples, including the interaction of HS and heparin-derived species with the chemokine IL-8, the growth factors FGF1 and FGF2, and the modulation of the activity of the enzyme heparanase by these species. Progresses in sequencing HS chains and reproducing them either by chemical synthesis or semi-synthesis, and in the elucidation of the 3D structure of oligosaccharide-protein complexes, are paving the way for rational approaches to the development of HS-inspired drugs in the field of inflammation and cancer, as well in other therapeutic fields.
Copyright © 2010 International Society of Matrix Biology. Published by Elsevier B.V. All rights reserved.
Figures
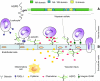
Fig. 1
(A) Model of a section of a HS chain (adapted from Murphy et al., 2004). (B) Multiple functions for HS during inflammatory reactions. HS participates in each of the major steps of leukocyte extravasation, such as facilitating L-selectin-dependent cell rolling, chemokine transport across the endothelium, and the chemokine presentation to leukocytes, which results in integrin activation (redrawn from Wang et al., 2005; Fig. 1 of Suppl. Inform.). ICAM: Intercellular adhesion molecule.
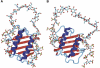
Fig. 2
Modeled complex of dimeric IL-8 and a heparin 24-mer composed of two hexasaccharides constituted of “fully sulfated” sequences connected by a nonsulfated N-acetylated, GlcA-containing dodecamer, before (A) and after (B) molecular dynamics simulation. In both binding sites, the interaction between protein and oligosaccharide focused on a disaccharide (Krieger et al., 2004).
Fig. 3
(A) Function of HS in growth factor (GF)-mediated signaling. (B) Activation of signaling by exogenous heparin (H) or a heparin-like HS fragment released by heparanase. The figure also shows two competing crystallographic models for the formation of ternary complexes involving FGF, FGFR, and a heparin dodecasaccharide. S–M: Schlessinger–Mohammadi symmetrical 2:2:2 complex for FGF2–FGFR1c; P–B: Pellegrini–Blundell asymmetrical 2:2:1 complex for FGF1–FGFR2c (structures redrawn from Mohammadi et al., 2005).
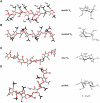
Fig. 4
Energy-minimized structures of heparin/HS S chains (A,B) and NA chains, these latter represented by the biosynthetic common precursor N-acetyl heparosan, C), and of one low-energy conformer of glycol-split N-acetyl heparin (D) compatible with experimental Nuclear Overhauser Effect values. Structures A and B (redrawn from Mulloy et al., 1993) illustrate the dramatic influence of changes in the conformation (from 1C4 to 2S0) of IdoA2OSO3 residues on spacing of sulfate groups along the chains (ANS and A6S = N-sulfate and 6-O-sulfate groups of GlcN residues; I2S = 2-O-sulfate group of IdoA residues). The rigid (4C1) conformation) of GlcA residues in N-acetyl heparosan does not involve significant changes from the chain conformation shown in C. Structures A–C are characterized by different dihedral angles between vicinal C–H bonds and distances between nonbonded atoms of the uronic acid residues. Structure D illustrates that glycol-splitting generates extra-degrees of rotational freedom (arrows), allowing abrupt kinks in the oligo/polysaccharide chains where one GlcA residue was periodate-oxidized and borohydride reduced. Similar conformations can be assumed by glycol-split, nonsulfated IdoA residues. (Panels A–C are from Casu, 2005; D from Vlodavsky et al., 2007.)
Fig. 5
(A) Details of the crystal structure of a 1:1 FGF2-heparin hexasaccharide complex, showing that the two 2-OSO3 residues select different conformations in binding to the growth factor (from Casu and Lindahl, 2001). (B) Partial solution structure of the complex of FGF2 and a synthetic tetrasaccharide, with both the sulfated iduronate residues in 1C4 conformation (Guglieri et al., 2008). Designation of residues as for Fig. 4; ΔU = 4,5-unsaturated uronic acid.
Similar articles
- Differential structural requirements of heparin and heparan sulfate proteoglycans that promote binding of basic fibroblast growth factor to its receptor.
Aviezer D, Levy E, Safran M, Svahn C, Buddecke E, Schmidt A, David G, Vlodavsky I, Yayon A. Aviezer D, et al. J Biol Chem. 1994 Jan 7;269(1):114-21. J Biol Chem. 1994. PMID: 8276782 - Heparanase and a synthetic peptide of heparan sulfate-interacting protein recognize common sites on cell surface and extracellular matrix heparan sulfate.
Marchetti D, Liu S, Spohn WC, Carson DD. Marchetti D, et al. J Biol Chem. 1997 Jun 20;272(25):15891-7. doi: 10.1074/jbc.272.25.15891. J Biol Chem. 1997. PMID: 9188488 - Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding.
Jemth P, Kreuger J, Kusche-Gullberg M, Sturiale L, Giménez-Gallego G, Lindahl U. Jemth P, et al. J Biol Chem. 2002 Aug 23;277(34):30567-73. doi: 10.1074/jbc.M203404200. Epub 2002 Jun 10. J Biol Chem. 2002. PMID: 12058038 - Heparin, heparan sulfate and heparanase in inflammatory reactions.
Li JP, Vlodavsky I. Li JP, et al. Thromb Haemost. 2009 Nov;102(5):823-8. doi: 10.1160/TH09-02-0091. Thromb Haemost. 2009. PMID: 19888515 Review. - Heparan sulfate and heparin interactions with proteins.
Meneghetti MC, Hughes AJ, Rudd TR, Nader HB, Powell AK, Yates EA, Lima MA. Meneghetti MC, et al. J R Soc Interface. 2015 Sep 6;12(110):0589. doi: 10.1098/rsif.2015.0589. J R Soc Interface. 2015. PMID: 26289657 Free PMC article. Review.
Cited by
- Sustained delivery of recombinant human bone morphogenetic protein-2 from perlecan domain I - functionalized electrospun poly (ε-caprolactone) scaffolds for bone regeneration.
Chiu YC, Fong EL, Grindel BJ, Kasper FK, Harrington DA, Farach-Carson MC. Chiu YC, et al. J Exp Orthop. 2016 Dec;3(1):25. doi: 10.1186/s40634-016-0057-1. Epub 2016 Oct 6. J Exp Orthop. 2016. PMID: 27714703 Free PMC article. - Syndecan-4 regulates extravillous trophoblast migration by coordinating protein kinase C activation.
Jeyarajah MJ, Jaju Bhattad G, Kops BF, Renaud SJ. Jeyarajah MJ, et al. Sci Rep. 2019 Jul 15;9(1):10175. doi: 10.1038/s41598-019-46599-6. Sci Rep. 2019. PMID: 31308409 Free PMC article. - Enzyme immobilization offers a robust tool to scale up the production of longer, diverse, natural glycosaminoglycan oligosaccharides.
Alabbas A, Desai UR. Alabbas A, et al. Glycobiology. 2020 Sep 28;30(10):768-773. doi: 10.1093/glycob/cwaa027. Glycobiology. 2020. PMID: 32193533 Free PMC article. - BuMPing iron with modified heparins.
Babitt JL, Lin HY. Babitt JL, et al. Blood. 2014 Mar 6;123(10):1440-1. doi: 10.1182/blood-2014-01-549519. Blood. 2014. PMID: 24627550 Free PMC article. - Modular synthesis of heparin-related tetra-, hexa- and octasaccharides with differential o-6 protections: programming for regiodefined 6-o-modifications.
Baráth M, Hansen SU, Dalton CE, Jayson GC, Miller GJ, Gardiner JM. Baráth M, et al. Molecules. 2015 Apr 9;20(4):6167-6180. doi: 10.3390/molecules20046167. Molecules. 2015. PMID: 25859776 Free PMC article.
References
- Ai X, Do A-T, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases. QSulf1 and QSulf2. J. Biol. Chem. 2006;281:4969–4976. - PubMed
- Angulo J, Ojeda R, de Paz J-L, Lucas R, Nieto PM, Lozano RM, Recondo-Horcajo M, Giménez-Gallego G, Martín-Lomas M. The activation of fibroblast growth factors (FGFs) by glycosaminoglycans: influence of the sulfation pattern on the biological activity of FGF1. ChemBioChem. 2004;5:55–61. - PubMed
- Asada M, Shinomiya M, Suzuki M, Honda E, Sugimoto R, Ikekita M, Imamura T. Glycosaminoglycan affinity of the complete fibroblast growth factor family. Biochim. Biophys. Acta. 2009;1790:40–48. - PubMed
- Ashikari-Hada S, Habuchi H, Karyia Y, Ytoh H, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J. Biol. Chem. 2004;279:12346–12354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical