Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells - PubMed (original) (raw)
. 2010 May 13;465(7295):175-81.
doi: 10.1038/nature09017. Epub 2010 Apr 25.
Affiliations
- PMID: 20418860
- PMCID: PMC3987905
- DOI: 10.1038/nature09017
Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells
Matthias Stadtfeld et al. Nature. 2010.
Abstract
Induced pluripotent stem cells (iPSCs) have been generated by enforced expression of defined sets of transcription factors in somatic cells. It remains controversial whether iPSCs are molecularly and functionally equivalent to blastocyst-derived embryonic stem (ES) cells. By comparing genetically identical mouse ES cells and iPSCs, we show here that their overall messenger RNA and microRNA expression patterns are indistinguishable with the exception of a few transcripts encoded within the imprinted Dlk1-Dio3 gene cluster on chromosome 12qF1, which were aberrantly silenced in most of the iPSC clones. Consistent with a developmental role of the Dlk1-Dio3 gene cluster, these iPSC clones contributed poorly to chimaeras and failed to support the development of entirely iPSC-derived animals ('all-iPSC mice'). In contrast, iPSC clones with normal expression of the Dlk1-Dio3 cluster contributed to high-grade chimaeras and generated viable all-iPSC mice. Notably, treatment of an iPSC clone that had silenced Dlk1-Dio3 with a histone deacetylase inhibitor reactivated the locus and rescued its ability to support full-term development of all-iPSC mice. Thus, the expression state of a single imprinted gene cluster seems to distinguish most murine iPSCs from ES cells and allows for the prospective identification of iPSC clones that have the full development potential of ES cells.
Figures
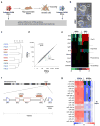
Figure 1. Aberrant silencing of the Dlk1-Dio3 gene cluster in mouse iPSCs
(a) Strategy for comparing genetically matched ESCs and iPSCs using “reprogrammable mice” harboring a doxycycline-inducible polycistronic reprogramming cassette (OKSM) in the Col1a1 (Collagen) locus. (b) Morphology of Collagen-OKSM ESCs and iPSCs. (c) Unsupervised clustering of four ESC and six iPSC lines based on microarray expression data. (d) Scatterplot of microarray data comparing iPSCs and ESCs with differentially expressed genes highlighted in green (2-fold, p0.05, t-test with Benjamini-Hochberg correction). (e) Heatmap showing relative expression levels of selected mRNAs in ESCs and iPSCs, covering in addition to Gtl2 and Rian other imprinted genes (Dlk1, Igf2r and H19) and pluripotency-associated transcripts (Nanog, Sox2 and Pou5f1). (f) Schematic representation of mouse chromosome 12 with position of the Dlk1-Dio3 gene cluster highlighted. Maternally-expressed and paternally-expressed transcripts are shown in red and blue, respectively. (g) Heatmap showing miRNAs that are differentially expressed between ESCs and iPSCs (2-fold, p0.01, t-test).
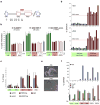
Figure 2. Full developmental potential of Gtl2on iPSCs
(a) Heatmap showing relative expression levels of Gtl2, Rian, other selected imprinted genes (Dlk1, H19 and Igf2r) and pluripotency-associated transcripts (Sox2 and Nanog) in ESCs and iPSCs derived from hematopoietic stem cells (HSC), granulocyte-macrophage progenitors (GMP), granulocytes (Gran), peritoneal fibroblasts (PF) and tail-tip fibroblasts (TTFs), isolated from three individual reprogrammable mice. Four iPSC clones expressing ESC-like levels of Gtl2 and Rian were identified (highlighted by asterisks) (for technical reasons, iPSC clone #18 could not be analyzed by microarray but instead was evaluated by qPCR. See Supplemental Figure 1b). (b) Strategy for assessing the developmental potential of iPSC clones by injection into diploid (2n) and tetraploid (4n) blastocysts to produce chimeric or all-iPSC mice, respectively. (c) Images of representative coat color chimeras with agouti indicating iPSC origin. (d) Coat color chimerism in mice derived from indicated Gtl2off (green diamonds), Gtl2on iPSC clones (red diamonds) and ESCs (open diamonds). (e) Statistical analysis of coat color chimerism in mice derived form Gtl2off and Gtl2on iPSC clones. (f) Image of two GFP+ all-iPSC pups (left panel) and two agouti all-iPSC mice (right). (g) Scatterplot showing intensity levels of all probesets covered by microarray analysis with those highlighted in green that were significantly different between 4n complementation-competent iPSCs (clones #19, #44, #47 and #49) and non-4n complementation-competent iPSCs (clones #18, #20, #45 and #48) (2-fold, p0.05, t-test with Benjamini-Hochberg correction).
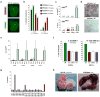
Figure 3. Epigenetic silencing of the Gtl2 locus in iPSCs
(a) Structure of the Dlk1-Dio3 locus with the position of the genomic regions analyzed by pyrosequencing indicated by black bars. (b) Degree of DNA methylation at IG-DMR and Gtl2 DMR in three Gtl2off iPSC clones (green bars), three Gtl2on iPSC clones (red bars), three ESCs clones (red open bars), as well as parental tail-tip fibroblasts (TTFs, grey bars). The methylation status of the other regions is shown in Supplemental Figure 5. (c) Prevalence of activation-associated (acH3, acH4 and H3K4me) and repression-associated (H3K27me) chromatin marks at the Gtl2 promoter in two Gtl2off iPSC clones, two Gtl2on iPSCs clones and ESCs. (d) Gtl2 expression levels as measured by qPCR in subclones derived from Gtl2off clone #45 and Gtl2on clone #49 in the absence (upper panel) or presence (lower panel) of doxycycline (dox). (e) Representative brightfield images of iPSCs culture in the absence or presence of all-trans retinoic acid (RA). (f) Expression levels of Gtl2, other imprinted genes (Igf2, Igf2r) and the pluripotency marker Pou5f1 in cells cultured with (+) or without (-) retinoic acid (RA). Note that the two Gtl2off clones fail to activate Gtl2, but show normal expression levels of the other imprinted genes.
Figure 4. Developmental defects in embryos derived from Gtl2off iPSCs
(a) Green fluorescence images of “all-iPSC” E11.5 embryos obtained with Gtl2on clone #47 (upper panel) and Gtl2off clone #48 (lower panel), both of which express EGFP from the ubiquitous ROSA26 locus. (b) Frequency of dead and living all-iPSC embryos obtained with two Gtl2on (red bars) and two Gtl2off (green bars) iPSC clones upon 4n blastocyst injection. Numbers of blastocysts transferred per clone and numbers of embryos recovered are indicated in brackets. (c) Expression of Glt2, Rian, Mirg and the paternally expressed gene Dlk1 in Gtl2off MEFs relative to Gtl2on MEFs (upper panel) as well as in Gtl2mKO MEFs relative to MEFs isolated from wildtype embryos (lower panel). (d) In situ hybridization against Gtl2 mRNA in MEFs derived from all-iPSC embryos generated with either Gtl2on clone #44 or Gtl2off clone #48. (e) Expression levels of Gtl2, Rian, Mirg and Dlk1 in the indicated tissues isolated from all-iPSC embryos made with Gtl2off iPSCs relative to the levels seen in tissues derived from Gtl2on iPSCs. (f) Degree of DNA methylation at the indicated regions in Gtl2off, Gtl2on, Gtl2mKO and wildtype MEFs. (g) Gtl2 expression levels in iPSC lines derived by subcloning Gtl2off clone #45 in the presence of valproic acid (VA). (h) Images of a fully developed stillborn pup (left) and a uterus filled with resorptions (right) derived after 4n blastocyst injections with either VA-10 or the parental iPSC clone #45, respectively.
Similar articles
- At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1.
Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM. Hagan JP, et al. PLoS One. 2009;4(2):e4352. doi: 10.1371/journal.pone.0004352. Epub 2009 Feb 5. PLoS One. 2009. PMID: 19194500 Free PMC article. - Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells.
Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, Park PJ, Hochedlinger K. Stadtfeld M, et al. Nat Genet. 2012 Mar 4;44(4):398-405, S1-2. doi: 10.1038/ng.1110. Nat Genet. 2012. PMID: 22387999 Free PMC article. - Hypermethylation and reduced expression of Gtl2, Rian and Mirg at the Dlk1-Dio3 imprinted locus as a marker for poor developmental potential of mouse embryonic stem cells.
Schacker M, Cheng YH, Eckersley-Maslin M, Snaith RM, Colledge WH. Schacker M, et al. Stem Cell Res. 2020 Oct;48:101931. doi: 10.1016/j.scr.2020.101931. Epub 2020 Jul 29. Stem Cell Res. 2020. PMID: 32822966 Free PMC article. - DLK1-DIO3 imprinted cluster in induced pluripotency: landscape in the mist.
Benetatos L, Vartholomatos G, Hatzimichael E. Benetatos L, et al. Cell Mol Life Sci. 2014 Nov;71(22):4421-30. doi: 10.1007/s00018-014-1698-9. Epub 2014 Aug 7. Cell Mol Life Sci. 2014. PMID: 25098353 Free PMC article. Review. - [Review on the genomic imprinting at the mammalian DLK1-DIO3 cluster.].
Zhao LX, Zhao GP, Zhou HM. Zhao LX, et al. Yi Chuan. 2010 Aug;32(8):769-78. doi: 10.3724/sp.j.1005.2010.00769. Yi Chuan. 2010. PMID: 20709673 Review. Chinese.
Cited by
- Manipulating cell fate through reprogramming: approaches and applications.
Yagi M, Horng JE, Hochedlinger K. Yagi M, et al. Development. 2024 Oct 1;151(19):dev203090. doi: 10.1242/dev.203090. Epub 2024 Sep 30. Development. 2024. PMID: 39348466 Review. - Knockdown H19 Accelerated iPSCs Reprogramming through Epigenetic Modifications and Mesenchymal-to-Epithelial Transition.
Sun R, Zhang X, Gong T, Zhang Y, Wang Q, He C, Ju J, Jin C, Ding W, Gao J, Shen J, Li Q, Shan Z. Sun R, et al. Biomolecules. 2024 Apr 23;14(5):509. doi: 10.3390/biom14050509. Biomolecules. 2024. PMID: 38785917 Free PMC article. - Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation.
Haubenreich C, Lenz M, Schuppert A, Peitz M, Koch P, Zenke M, Brüstle O. Haubenreich C, et al. Int J Mol Sci. 2024 Mar 12;25(6):3214. doi: 10.3390/ijms25063214. Int J Mol Sci. 2024. PMID: 38542188 Free PMC article. - Therapeutic potential of mesenchymal stem cells from human iPSC-derived teratomas for osteochondral defect regeneration.
Kim J, Kim JS, Kim D, Bello AB, Kim BJ, Cha BH, Lee SH. Kim J, et al. Bioeng Transl Med. 2023 Nov 29;9(2):e10629. doi: 10.1002/btm2.10629. eCollection 2024 Mar. Bioeng Transl Med. 2023. PMID: 38435815 Free PMC article. - Metabolic control of induced pluripotency.
Sinenko SA, Tomilin AN. Sinenko SA, et al. Front Cell Dev Biol. 2024 Jan 11;11:1328522. doi: 10.3389/fcell.2023.1328522. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38274274 Free PMC article. Review.
References
- Takahashi K, Yamanaka S. Cell. 2006;126(4):663. - PubMed
- Zhao XY, Li W, Lv Z, et al. Nature. 2009
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources