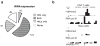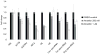Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes - PubMed (original) (raw)
Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes
Artem Barski et al. Nat Struct Mol Biol. 2010 May.
Abstract
Epigenetic control is an important aspect of gene regulation. Despite detailed understanding of protein-coding gene expression, the transcription of noncoding RNA genes by RNA polymerase III (Pol III) is less well characterized. Here we profile the epigenetic features of Pol III target genes throughout the human genome. This reveals that the chromatin landscape of Pol III-transcribed genes resembles that of Pol II templates in many ways, although there are also clear differences. Our analysis also uncovered an entirely unexpected phenomenon: namely, that Pol II is present at the majority of genomic loci that are bound by Pol III.
Figures
Figure 1
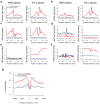
Chromatin environment of pol III genes. (a,b) Expressed and silent copies of tRNA—Leu-TAA, (c) U6 RNA, (d) 7SK RNA, (e) HVG1 RNA, (f) RNase MRP RNA. The positions of the non-coding RNA genes are indicated at the bottom of each panel. Bars show number of ChIP-Seq tags in 200bp windows and RNA-Seq tags in 20bp windows.
Figure 2
Pol III target genes are associated with both similar and distinct chromatin features to pol II target genes. Average ChIP-Seq read density (reads per 100 bp) profiles at tRNA (left) and pol II (right) genes are plotted for (a) H3K4me3, (b) H3K9ac, (c) H2A.Z, (d) H3K27me3, (e)H3K79me2, (f) H3K36me3. Genes were imperfectly classified as expressed or silent based on presence of pol III for tRNA genes or pol II for pol II genes. Area surrounding TSS (TES for H3K36me3)- transcription start (end) site is shown. Profiles for other modifications are shown in Supplementary Fig. 1. (g) Histone H3 ChIP-Seq read densities surrounding the TSSs of expressed (red) and silent (blue) tRNA genes.
Figure 3
Tissue specific expression of tRNA genes. (a) Pie chart shows number of tRNAs expressed in HeLa cells, CD4+ T cells, both or neither. (b) Chromatin environment of tRNA—Glu-TTC in CD4+ T and HeLa cells.
Figure 4
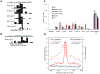
Novel pol III binding sites. (a,b,c) Novel pol III sites were found in the vicinity of known pol II promoters (a,b) as well as in gene free areas. Presence of total RNA-Seq signals suggests that the binding of polymerase at these loci is productive. Absence of corresponding polyA RNA-seq signal in b and c suggests that these loci are transcribed by pol III rather than pol II. (d) TFIIIB and TFIIIC can be found at the majority of the novel pol III loci.
Figure 5
Pol II is present at pol III-transcribed genes. (a) Various pol II isoforms are present at tRNASer-TGA in human CD4+ Tcells, (b) pol II is present at two tRNAs in Drosophila S2 cells. (c) Recruitment of pol II and its factors to various Pol III-transcribed RNA genes. (d) ChIP-Seq tag density profiles of pol II and III in the vicinity of silent and expressed tRNA genes.
Figure 6
Levels of 5S rRNA and pre-tRNAs are decreased by 4 hour treatment with alpha-amanitin oleate. qPCR results show that levels of both pol II (ACTB, GAPDH and IRF2) and some pol III genes (5S rRNA and pre-tRNAs) are decreased as a result of short term treatment with pol II inhibitor alpha-amanitin oleate. Mean +/−SD are shown, n=3.
Similar articles
- Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors.
Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Oler AJ, et al. Nat Struct Mol Biol. 2010 May;17(5):620-8. doi: 10.1038/nsmb.1801. Epub 2010 Apr 25. Nat Struct Mol Biol. 2010. PMID: 20418882 Free PMC article. - Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells.
Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Moqtaderi Z, et al. Nat Struct Mol Biol. 2010 May;17(5):635-40. doi: 10.1038/nsmb.1794. Epub 2010 Apr 25. Nat Struct Mol Biol. 2010. PMID: 20418883 Free PMC article. - Compromised RNA polymerase III complex assembly leads to local alterations of intergenic RNA polymerase II transcription in Saccharomyces cerevisiae.
Wang Q, Nowak CM, Korde A, Oh DH, Dassanayake M, Donze D. Wang Q, et al. BMC Biol. 2014 Oct 28;12:89. doi: 10.1186/s12915-014-0089-x. BMC Biol. 2014. PMID: 25348158 Free PMC article. - Epigenetic regulation of noncoding RNA transcription by mammalian RNA polymerase III.
Park JL, Lee YS, Kunkeaw N, Kim SY, Kim IH, Lee YS. Park JL, et al. Epigenomics. 2017 Feb;9(2):171-187. doi: 10.2217/epi-2016-0108. Epigenomics. 2017. PMID: 28112569 Review. - Epigenetic regulation of transcription by RNA polymerase III.
Bhargava P. Bhargava P. Biochim Biophys Acta. 2013 Oct;1829(10):1015-25. doi: 10.1016/j.bbagrm.2013.05.005. Epub 2013 Jun 1. Biochim Biophys Acta. 2013. PMID: 23732820 Review.
Cited by
- Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators.
James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. James Faresse N, et al. PLoS Genet. 2012;8(11):e1003028. doi: 10.1371/journal.pgen.1003028. Epub 2012 Nov 15. PLoS Genet. 2012. PMID: 23166507 Free PMC article. - The emerging role of lncRNAs in the regulation of cancer stem cells.
Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. Castro-Oropeza R, et al. Cell Oncol (Dordr). 2018 Dec;41(6):585-603. doi: 10.1007/s13402-018-0406-4. Epub 2018 Sep 14. Cell Oncol (Dordr). 2018. PMID: 30218296 Review. - Y RNAs are conserved endogenous RIG-I ligands across RNA virus infection and are targeted by HIV-1.
Vabret N, Najburg V, Solovyov A, Gopal R, McClain C, Šulc P, Balan S, Rahou Y, Beauclair G, Chazal M, Varet H, Legendre R, Sismeiro O, Sanchez David RY, Chauveau L, Jouvenet N, Markowitz M, van der Werf S, Schwartz O, Tangy F, Bhardwaj N, Greenbaum BD, Komarova AV. Vabret N, et al. iScience. 2022 Jun 11;25(7):104599. doi: 10.1016/j.isci.2022.104599. eCollection 2022 Jul 15. iScience. 2022. PMID: 35789859 Free PMC article. - Gene-specific RNA polymerase II phosphorylation and the CTD code.
Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Kim H, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1279-86. doi: 10.1038/nsmb.1913. Epub 2010 Sep 12. Nat Struct Mol Biol. 2010. PMID: 20835241 Free PMC article. - Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription.
Carvalho S, Raposo AC, Martins FB, Grosso AR, Sridhara SC, Rino J, Carmo-Fonseca M, de Almeida SF. Carvalho S, et al. Nucleic Acids Res. 2013 Mar 1;41(5):2881-93. doi: 10.1093/nar/gks1472. Epub 2013 Jan 15. Nucleic Acids Res. 2013. PMID: 23325844 Free PMC article.
References
- Dieci G, et al. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. - PubMed
- Yang Z, et al. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. - PubMed
- Nguyen VT, et al. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. - PubMed
- Mossink MH, van Zon A, Scheper RJ, Sonneveld P, Wiemer EAC. Vaults: a ribonucleoprotein particle involved in drug resistance? Oncogene. 2003;22:7458–7467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K22 HL098691/HL/NHLBI NIH HHS/United States
- Z01 HL005801/ImNIH/Intramural NIH HHS/United States
- 1K22HL098691-01/HL/NHLBI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases