Metagenomic sequencing of an in vitro-simulated microbial community - PubMed (original) (raw)
Metagenomic sequencing of an in vitro-simulated microbial community
Jenna L Morgan et al. PLoS One. 2010.
Abstract
Background: Microbial life dominates the earth, but many species are difficult or even impossible to study under laboratory conditions. Sequencing DNA directly from the environment, a technique commonly referred to as metagenomics, is an important tool for cataloging microbial life. This culture-independent approach involves collecting samples that include microbes in them, extracting DNA from the samples, and sequencing the DNA. A sample may contain many different microorganisms, macroorganisms, and even free-floating environmental DNA. A fundamental challenge in metagenomics has been estimating the abundance of organisms in a sample based on the frequency with which the organism's DNA was observed in reads generated via DNA sequencing.
Methodology/principal findings: We created mixtures of ten microbial species for which genome sequences are known. Each mixture contained an equal number of cells of each species. We then extracted DNA from the mixtures, sequenced the DNA, and measured the frequency with which genomic regions from each organism was observed in the sequenced DNA. We found that the observed frequency of reads mapping to each organism did not reflect the equal numbers of cells that were known to be included in each mixture. The relative organism abundances varied significantly depending on the DNA extraction and sequencing protocol utilized.
Conclusions/significance: We describe a new data resource for measuring the accuracy of metagenomic binning methods, created by in vitro-simulation of a metagenomic community. Our in vitro simulation can be used to complement previous in silico benchmark studies. In constructing a synthetic community and sequencing its metagenome, we encountered several sources of observation bias that likely affect most metagenomic experiments to date and present challenges for comparative metagenomic studies. DNA preparation methods have a particularly profound effect in our study, implying that samples prepared with different protocols are not suitable for comparative metagenomics.
Conflict of interest statement
Competing Interests: Jonathan Eisen is an associate with PLoS as Editor-in-Chief of PLoS Biology.
Figures
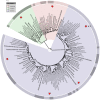
Figure 1. Phylogenetic distribution of organisms selected for the metagenomic simulation.
A phylogenetic tree of three domains with representative groups is shown. Organisms used in this study are indicated by *. The organisms used represent all known domains of life, include four bacterial phyla, a variety of genome sizes, GC compositions, and cell wall types. Large font size indicates clades where multiple isolate genomes have been collapsed into a single leaf node.
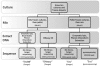
Figure 2. Outline of the steps involved in the creation and sequencing of the simulated metagenomic samples.
Figure 3. Predicted and observed frequencies of sequence reads from each organism.
The fraction of reads assigned to organisms for each sample preparation method is shown at top. The fraction expected given the measured quantities of mixed DNA from each organism assuming unbiased library prep and sequencing is given as “DNA quantification”, and the fraction of reads predicted based on cell count and genome size is given as “cc*gs prediction.” Sampling error was estimated assuming a multinomial distribution (not shown) and indicated that estimates of relative abundance are accurate +/−5% for dominant organisms given the number of Sanger reads obtained, and +/−1% for pyrosequencing reads. Note that the top two bars labeled Enz+Pyrosequencing and Enz+Sanger offer a comparison of Sanger and pyrosequencing technology on the same extracted DNA.
Figure 4. Additional sequence data for three of the simulated metagenomes.
Bars represent the observed frequency of organisms in sequenced metagenomes. We constructed and sequenced metagenomes according to the Enz, EnzBB, and DNeasy protocols using the long term frozen isolate culture stocks with glycerol and without glycerol. Reads were mapped to reference genomes as described in Methods. The additional metagenomes show some differences to each of the original libraries. Such differences might be caused by variation across DNA preparations and sequencing runs, age of the frozen samples, or other factors. The libraries constructed using the DNeasy Kit produced the most consistent results.
Similar articles
- A novel abundance-based algorithm for binning metagenomic sequences using l-tuples.
Wu YW, Ye Y. Wu YW, et al. J Comput Biol. 2011 Mar;18(3):523-34. doi: 10.1089/cmb.2010.0245. J Comput Biol. 2011. PMID: 21385052 Free PMC article. - MBMC: An Effective Markov Chain Approach for Binning Metagenomic Reads from Environmental Shotgun Sequencing Projects.
Wang Y, Hu H, Li X. Wang Y, et al. OMICS. 2016 Aug;20(8):470-9. doi: 10.1089/omi.2016.0081. Epub 2016 Jul 22. OMICS. 2016. PMID: 27447888 Free PMC article. - Estimating DNA coverage and abundance in metagenomes using a gamma approximation.
Hooper SD, Dalevi D, Pati A, Mavromatis K, Ivanova NN, Kyrpides NC. Hooper SD, et al. Bioinformatics. 2010 Feb 1;26(3):295-301. doi: 10.1093/bioinformatics/btp687. Epub 2009 Dec 14. Bioinformatics. 2010. PMID: 20008478 Free PMC article. - From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems.
Garza DR, Dutilh BE. Garza DR, et al. Cell Mol Life Sci. 2015 Nov;72(22):4287-308. doi: 10.1007/s00018-015-2004-1. Epub 2015 Aug 9. Cell Mol Life Sci. 2015. PMID: 26254872 Free PMC article. Review. - From bacterial to microbial ecosystems (metagenomics).
Williamson SJ, Yooseph S. Williamson SJ, et al. Methods Mol Biol. 2012;804:35-55. doi: 10.1007/978-1-61779-361-5_3. Methods Mol Biol. 2012. PMID: 22144147 Review.
Cited by
- How Do the Players Play? A Post-Genomic Analysis Paradigm to Understand Aquatic Ecosystem Processes.
Reid T, Bergsveinson J. Reid T, et al. Front Mol Biosci. 2021 May 7;8:662888. doi: 10.3389/fmolb.2021.662888. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026835 Free PMC article. - Inferring dynamic signatures of microbes in complex host ecosystems.
Gerber GK, Onderdonk AB, Bry L. Gerber GK, et al. PLoS Comput Biol. 2012;8(8):e1002624. doi: 10.1371/journal.pcbi.1002624. Epub 2012 Aug 2. PLoS Comput Biol. 2012. PMID: 22876171 Free PMC article. - Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure.
Narrowe AB, Albuthi-Lantz M, Smith EP, Bower KJ, Roane TM, Vajda AM, Miller CS. Narrowe AB, et al. Microbiome. 2015 Mar 3;3:6. doi: 10.1186/s40168-015-0069-6. eCollection 2015. Microbiome. 2015. PMID: 25815185 Free PMC article. - Molecular methods for pathogen and microbial community detection and characterization: current and potential application in diagnostic microbiology.
Sibley CD, Peirano G, Church DL. Sibley CD, et al. Infect Genet Evol. 2012 Apr;12(3):505-21. doi: 10.1016/j.meegid.2012.01.011. Epub 2012 Feb 9. Infect Genet Evol. 2012. PMID: 22342514 Free PMC article. Review. - An Optimized Active Sampling Procedure for Aerobiological DNA Studies.
Basapathi Raghavendra J, Mathanlal T, Zorzano MP, Martin-Torres J. Basapathi Raghavendra J, et al. Sensors (Basel). 2023 Mar 5;23(5):2836. doi: 10.3390/s23052836. Sensors (Basel). 2023. PMID: 36905039 Free PMC article.
References
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. - PubMed
- Blow N. Metagenomics: exploring unseen communities. Nature. 2008;453:687–690. - PubMed
- Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3:470–478. - PubMed