Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores - PubMed (original) (raw)
Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores
Takahito Yoshida et al. PLoS Genet. 2010.
Abstract
Actin-related proteins are ubiquitous components of chromatin remodelers and are conserved from yeast to man. We have examined the role of the budding yeast actin-related protein Arp6 in gene expression, both as a component of the SWR1 complex (SWR-C) and in its absence. We mapped Arp6 binding sites along four yeast chromosomes using chromatin immunoprecipitation from wild-type and swr1 deleted (swr1Delta) cells. We find that a majority of Arp6 binding sites coincide with binding sites of Swr1, the catalytic subunit of SWR-C, and with the histone H2A variant Htz1 (H2A.Z) deposited by SWR-C. However, Arp6 binding detected at centromeres, the promoters of ribosomal protein (RP) genes, and some telomeres is independent of Swr1 and Htz1 deposition. Given that RP genes and telomeres both show association with the nuclear periphery, we monitored the ability of Arp6 to mediate the localization of chromatin to nuclear pores. Arp6 binding is sufficient to shift a randomly positioned locus to nuclear periphery, even in a swr1Delta strain. Arp6 is also necessary for the pore association of its targeted RP promoters possibly through cell cycle-dependent factors. Loss of Arp6, but not Htz1, leads to an up-regulation of these RP genes. In contrast, the pore-association of GAL1 correlates with Htz1 deposition, and loss of Arp6 reduces both GAL1 activation and peripheral localization. We conclude that Arp6 functions both together with the nucleosome remodeler Swr1 and also without it, to mediate Htz1-dependent and Htz1-independent binding of chromatin domains to nuclear pores. This association is shown to have modulating effects on gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
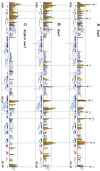
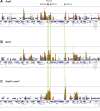
Figure 1. Localization of Arp6 and Swr1 on chromosome 6R.
Vertical bars represent the binding ratio of proteins at each locus. Filled bars with yellow and black were determined to be significantly positive, for loci covering 300-bp or 100-bp regions, respectively. Open bars were not significantly positive. The scale of the vertical axis is log2 and the upper and lower horizontal lines represent 1.0 and -1.0, respectively. The central horizontal axis shows kilobase units. (A) Localization of Arp6-FLAG in SWR1 wild-type cells. (B) Localization of Swr1-FLAG. (C) Localization of Arp6-FLAG in swr1 cells. Vertical arrows (I to X) in A and vertical green lines represent the position of the highest ten clusters consisting of at least two continuous binding loci in both Arp6-and Swr1-FLAG ChIP.
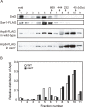
Figure 2. Sizing column Superose6 analysis of Arp6-complexes.
(A) Extracts from cells expressing Arp6-FLAG under wild-type and swr1 background were fractionated on a Superose6 column, and Arp6-FLAG in the fractions was detected on a Western blot with an anti-FLAG antibody (two bottom panels). The extract from cells expressing Swr1-FLAG was fractionated as well, and endogenous Snf2 and Swr1-FLAG were detected with an anti-Snf2 antibody and an anti-FLAG antibody, respectively (two top panels). The numbers of fractions and the positions of molecular mass standards are shown over the panels. (B) The intensity of Arp6-FLAG in the fractions were quantified, and relative distributions of Arp6 in the fractions of wild-type (WT) and swr1 cells were shown.
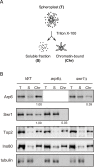
Figure 3. Arp6 partitions between soluble and insoluble chromatin fractions.
(A) The fractionation protocol is shown. Yeast spheroplasts from appropriate strains were lysed with Triton-X100. A gentle centrifugation step separates a supernatant containing the bulk of cellular proteins from a chromatin pellet. (B) Wild-type, swr1, and arp6 cells were subjected to the fractionation protocol described in (A), and the spheroplast (T), soluble fraction (S), and chromatin-bound (Chr) samples were probed using Western blot for Arp6-FLAG, Swr1-FLAG, topoisomerase II (Top2), the enzymatic component of the INO80 chromatin remodeling complexes (Ino80-MYC), and the soluble non-chromatin protein, tubulin. Numbers under panels show the ratios of chromatin-bound Arp6 and Swr1 in the mutants relative to WT. Their intensities were normalized with chromatin-preparation efficiencies obtained by quantification of the Western blot for Ino80.
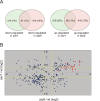
Figure 4. Transcript analysis in swr1 and arp6 mutant cells.
Microarray analysis was repeated at least three times for swr1Δ and arp6Δ strain, and the statistical differences were determined using a t-test. p<0.05 was considered significant. (A) The Venn diagrams illustrate the degree of overlap between genes whose RNA levels were changed by 1.25-fold in arp6 and swr1 mutants. Numbers correspond to misregulated genes in the total genes. Relative numbers of the misregulated genes (%/total number of yeast gene) are indicated in parentheses. (B) Genes whose transcriptional changes were statistically significant (p<0.05) both in arp6 and swr1 cells were plotted according to their log2 ratios. The yellow lines show the 1.25-fold changes. The red diamonds represents RP genes.
Figure 5. Swr1-independent binding of Arp6 to RP genes.
Vertical bars represent the binding ratio of proteins in each locus. The binding of Arp6-FLAG (top), Swr1-FLAG (middle), and Arp6-FLAG in swr1 cells (bottom) in the region 266K-353K of Chr 4L were compared. The positions of the RP genes (RPS16B, RPL13A, RPP1A, and RPL31A) in the region are shown with arrows and green lines. Red asterisks indicate those Arp6-gene promoter bindings which disappeared in the absence of Swr1.
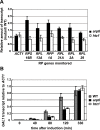
Figure 6. Quantitative analysis of transcripts in cells lacking Arp6 (arp6) and H2A.Z (htz1) using RT–PCR.
(A) The same amount of total RNA from wild-type, arp6, and htz1 cells was analyzed using quantitative RT–PCR by using primer sets specific for each of the RP genes. The ACT1 gene was analyzed as a control. The relative amount of the transcript of the genes in arp6 and htz1 to wild-type is shown by filled and open bars, respectively. The data points represent the mean ± SD for at least three experiments. (B) Total RNA was prepared from wild-type, arp6, and htz1 cells (gray, filled, and open bars, respectively) at the indicated times after induction of GAL1 expression in galactose. The amounts of GAL1 transcript in these strains were analyzed using quantitative RT–PCR, and shown as relative amounts compared to ACT1 transcript. The data points represent the mean ± SD for at least three experiments.
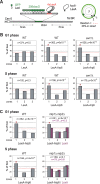
Figure 7. Perinuclear anchoring activity of Arp6.
(A) The ability of a LexA fusion to relocate the lacO-tagged PES4 locus bearing LexA binding sites was tested using a strain GA-1461. PES4 is located 70 kb and 50 kb from Tel6R and Cen6, respectively. The lacO array was visualized by binding a GFP-LacI fusion, and the nuclear envelope is visualized through a Nup49-GFP fusion. The focal plane in which the GFP spot was brightest was used to monitor distances, which were reported as a ratio to nuclear diameter. These values were binned into one of three concentric zones of equal surface and are presented as percentage of total spots scored. The position of PES4 was mapped in wild-type (WT) and swr1 cells expressing LexA alone or a LexA-Arp6 fusion. (B) Arp6 can relocalize an internal chromatin locus (PES4) to the nuclear periphery. Cells were classified as G1 (unbudded) or S phase (budded with spherical nucleus). (C) Mlp1/Mlp2 is required for the perinuclear anchoring by Arp6 in S-phase cells. The position of the lacO arrays was scored on PES4::lacO tagged cells expressing LexA (purple) or LexA-Arp6 (grey). The bar graphs show the percentage of spots (y-axis) per zone (x-axis) for G1-phase (upper panels) and S-phase (lower panels) cells. The number of cells analyzed (n) and the confidence values (p) for the χ2 analysis between random and test distributions are indicated. The red dashed horizontal line at 33% indicates a random distribution, and zone 1 distributions that are significantly different from random are indicated by an asterisk (p<0.05).
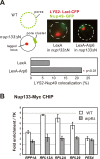
Figure 8. Involvement of Arp6 in intranuclear organization through the NPC.
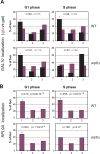
(A) The positions of _lacO_-tagged LYS2 (red) and of CFP-Nup49 (green) were observed in a nup133ΔN background, in which nuclear pores cluster on one side of the nucleus. Bar graphs represent the percentage of complete red-green signal overlap counted in cells expressing LexA alone or a LexA-Arp6 fusion. The confidence values (p) for the χ2 analysis between them is indicated. The predicted colocalization for a randomly positioned locus is 9% . (B) Requirement of Arp6 for the interaction of RP genes with the NPC. The association of Nup133-Myc with RP genes, RPP1A, RPL13A, RPL2A, and RPL29, was quantified using ChIP analysis combined with quantitative PCR in wild-type (WT) and arp6 cells, and is plotted over a background control of the TK gene . The PES4 locus was analyzed as a control. The data points represent the mean ± SD for at least three experiments.
Figure 9. Arp6 is required for the peripheral association of galactose-induced GAL10 and constitutively expressed RP gene RPL9A.
(A) The GAL1-GAL10 locus was tagged by inserting 256 lac operators in a haploid wild-type or arp6 deletion strain bearing GFP-lacI and Nup49-GFP fusions (wild-type; GA-4098, arp6; GA-6024) . The position of the lacO arrays relative to the nuclear envelope was scored on images take of living cells growth either on glucose (purple) or after 2 hours of gene induction on 2% galactose (black). Three zone scoring was carried out as in Figure 7. The number of cells analyzed for each stage of the cell cycle are indicated, and the confidence values (p) for the χ2 analysis between random and test distributions on galactose are: wild-type (G1, p = 4×10−4; S, p = 6×10−4) and arp6 (G1, p = 2.7×10−8; S, p = 0.44) none of the values on glucose are significantly different from random (p>0.05). The G1-S differences on galactose are not significant in WT, but are in the arp6 mutant (p = 0.024). Note that in this analysis we omitted the rare, very small budded cells. (B) The RPL9A locus was tagged by inserting 256 lac operators in a haploid wild-type or arp6 deletion strain bearing GFP-lacI and Nup49-GFP fusions (wild-type; GA-3635, arp6; GA-5132) . The position of the lacO arrays relative to the nuclear envelope was scored as in Figure 7 on image stacks taken on living cells grown on SC. Symbols and quantitation are as in A. RPL9A locus p values for test vs random distributions are: wild-type (G1, p = 9.8×10−13; S, p = 4.9×10−12) and arp6 (G1, p = 7.6×10−9; S, p = 0.07). The G1 vs S distributions in wild-type are not significantly different (p = 0.33) while in arp6 cells the difference is significant (p = 0.028). Whereas values for Zone 1 in wild-type vs arp6 cells in G1 are not significantly different (p = 0.82), the difference in mid-to-late S phase cells is (p = 0.0018). An asterisk indicates that values that have a nonrandom distribution (p<0.05). The number of cells analyzed (n) and the confidence values (p) for the χ2 analysis between random and test distributions are indicated.
Similar articles
- Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition.
Potok ME, Wang Y, Xu L, Zhong Z, Liu W, Feng S, Naranbaatar B, Rayatpisheh S, Wang Z, Wohlschlegel JA, Ausin I, Jacobsen SE. Potok ME, et al. Nat Commun. 2019 Jul 26;10(1):3352. doi: 10.1038/s41467-019-11291-w. Nat Commun. 2019. PMID: 31350403 Free PMC article. - Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z.
Deal RB, Topp CN, McKinney EC, Meagher RB. Deal RB, et al. Plant Cell. 2007 Jan;19(1):74-83. doi: 10.1105/tpc.106.048447. Epub 2007 Jan 12. Plant Cell. 2007. PMID: 17220196 Free PMC article. - Arabidopsis cryptochrome 1 controls photomorphogenesis through regulation of H2A.Z deposition.
Mao Z, Wei X, Li L, Xu P, Zhang J, Wang W, Guo T, Kou S, Wang W, Miao L, Cao X, Zhao J, Yang G, Zhang S, Lian H, Yang HQ. Mao Z, et al. Plant Cell. 2021 Jul 19;33(6):1961-1979. doi: 10.1093/plcell/koab091. Plant Cell. 2021. PMID: 33768238 Free PMC article. - The specificity of H2A.Z occupancy in the yeast genome and its relationship to transcription.
Iyer VR. Iyer VR. Curr Genet. 2020 Oct;66(5):939-944. doi: 10.1007/s00294-020-01087-7. Epub 2020 Jun 14. Curr Genet. 2020. PMID: 32537667 Review. - The beauty of being a variant: H2A.Z and the SWR1 complex in plants.
March-Díaz R, Reyes JC. March-Díaz R, et al. Mol Plant. 2009 Jul;2(4):565-577. doi: 10.1093/mp/ssp019. Epub 2009 Mar 31. Mol Plant. 2009. PMID: 19825639 Review.
Cited by
- PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL.
Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, Gasser SM. Horigome C, et al. Genes Dev. 2016 Apr 15;30(8):931-45. doi: 10.1101/gad.277665.116. Epub 2016 Apr 7. Genes Dev. 2016. PMID: 27056668 Free PMC article. - A human XPC protein interactome--a resource.
Lubin A, Zhang L, Chen H, White VM, Gong F. Lubin A, et al. Int J Mol Sci. 2013 Dec 23;15(1):141-58. doi: 10.3390/ijms15010141. Int J Mol Sci. 2013. PMID: 24366067 Free PMC article. - The INO80 Complex Requires the Arp5-Ies6 Subcomplex for Chromatin Remodeling and Metabolic Regulation.
Yao W, King DA, Beckwith SL, Gowans GJ, Yen K, Zhou C, Morrison AJ. Yao W, et al. Mol Cell Biol. 2016 Jan 11;36(6):979-91. doi: 10.1128/MCB.00801-15. Mol Cell Biol. 2016. PMID: 26755556 Free PMC article. - The transcriptional state and chromatin landscape of cichlid jaw shape variation across species and environments.
Tetrault E, Swenson J, Aaronson B, Marcho C, Albertson RC. Tetrault E, et al. Mol Ecol. 2023 Jul;32(14):3922-3941. doi: 10.1111/mec.16975. Epub 2023 May 9. Mol Ecol. 2023. PMID: 37160741 Free PMC article. - Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans.
Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Rohner S, et al. J Cell Biol. 2013 Mar 4;200(5):589-604. doi: 10.1083/jcb.201207024. J Cell Biol. 2013. PMID: 23460676 Free PMC article.
References
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. - PubMed
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. - PubMed
- Akhtar A, Gasser S. The nuclear envelope and transcriptional control. Nature Rev. 2007;8:507–517. - PubMed
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials