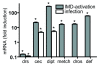Chronic activation of the epithelial immune system of the fruit fly's salivary glands has a negative effect on organismal growth and induces a peculiar set of target genes - PubMed (original) (raw)
Chronic activation of the epithelial immune system of the fruit fly's salivary glands has a negative effect on organismal growth and induces a peculiar set of target genes
Ahmed Abdelsadik et al. BMC Genomics. 2010.
Abstract
Background: Epithelial and especially mucosal immunity represents the first line of defence against the plethora of potential pathogens trying to invade via the gastrointestinal tract. The salivary glands of the fruit fly are an indispensable part of the gastrointestinal tract, but their contribution to the mucosal immunity has almost completely been neglected. Our major goal was to elucidate if the fly's salivary glands are able to mount an immune response and what the major characteristics of this immune response are.
Results: Ectopic activation of the IMD-pathway within the salivary gland cells is able to induce an immune response, indicating that the salivary glands are indeed immune competent. This reaction is characterized by the concurrent expression of numerous antimicrobial peptide genes. In addition, ectopic activation of the salivary gland's immune response induces morphological changes such as dwarfism throughout all developmental stages and a significantly decreased length of the salivary glands themselves. DNA-microarray analyses of the reaction revealed a complex pattern of up- and downregulated genes. Gene ontology analyses of regulated genes revealed a significant increase in genes associated with ribosomal and proteasomal function. On the other hand, genes coding for peptide receptors and some potassium channels are downregulated. In addition, the comparison of the transcriptional events induced following IMD-activation in the trachea and the salivary glands shows also only a small overlap, indicating that the general IMD-activated core transcriptome is rather small and that the tissue specific component of this response is dominating. Among the regulated genes, those that code for signaling associated protease activity are significantly modulated.
Conclusions: The salivary glands are immune-competent and they contribute to the overall intestinal immune system. Although they produce antimicrobial peptides, their overall response is highly tissue-specific. Our analysis indicates that chronic activation of the salivary gland's immune system is costly, as it induces severe reduction in growth throughout development. The IMD-regulated increase in expression levels of the fly's presenilin representatives opens the opportunity to use the salivary glands for studying the physiological and pathophysiological role of these genes in a simple but functional environment.
Figures
Figure 1
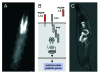
Using the Gal4/UAS-system, activation of the salivary glands can be achieved by genetic means. Proteins of the salivary gland secretion family are believed to be restricted to the salivary glands. Crossing the sgs3::gal4 line with a UAS::gfp responder line revealed that expression is indeed restricted to the salivary glands (A). Based on the UAS/Gal4 system for ectopic activation, the IMD-pathway, which is instrumental in epithelial immunity, can be activated by targeted overexpression of peptidoglycan recognition receptors. The membrane bound PGRP-LC activates in a cell-autonomous way, whereas the soluble PGRP-LE activates in an organ-wide, systemic way (B). If the pgrp-le gene is expressed in the salivary glands only, the antimicrobial peptide gene drosomycin (here used as a reporter for immune system activity and visualized by drosP::gfp) is expressed in defined parts of the intestine only (but not on the salivary glands) (C).
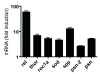
Figure 2
Infection and ectopic activation of the IMD-pathway induces expression of antimicrobial peptides in the salivary glands. Oral infection of early 3rd instar with the insect pathogen Erwinia carotovora was used to monitor transcriptional changes in 3 selected antimicrobial peptide genes, drosomycin (drs), cecropin (cec) and diptericin (dipt)(white bars). Ectopic overexpression of the prgp-lc gene in the larval salivary glands was achieved using the Gal4/UAS system (sgs3::gal4 X UAS::pgrp-lc). Salivary glands of early 3rd instar larvae were isolated from these crossing and a parental line (responder line) used for control. Quantitative real-time PCR was performed with oligonucleotides comprising approximately 150 bp of the corresponding antimicrobial peptide genes; drosomycin (drs), cecropin (cec), diptericin (dipt), metchnikowin (metch), drosomycin (dros) and defensin (def) (grey bars). Results are the mean of at least 3 experiments performed in triplicate. Controls are set to 1. Statistically significant differences (compared with controls are marked by an asterik (p < 0.05).
Figure 3
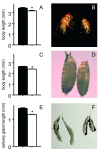
Chronic ectopic activation of the IMD-pathway in the salivary glands only had a significant effect on growth and body length of the animals. Adult males, where PGRP-LC is ectopically expressed in the salivary gland have an approximately 10% reduced body length if compared with the parental line (A, B). A similar reduction can be seen in the pupal length of both types of animals (C, D). Regarding the lengths of isolated salivary glands from the corresponding animals, these differences are even more pronounced (E, F). For A, C, E, median values of at least 20 measurements +/- SEM. Statistically significant differences are marked by an asterik (p < 0.05).
Figure 4
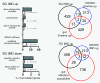
DNA-microarray analyses of the transcriptional changes within the salivary glands that occur following chronic activation of the IMD-pathway in these cells only. Cohorts of genes were identified that are either upregulated (additional file 3) or downregulated (additional file 4). Only those genes are included, where these differences are greater than twofold and the differences are statistically significant (evaluated with the SAM software package). Gene ontology analyses were performed with these cohorts of genes and compared to the complete set of Drosophila genes using the Fatigo software tool [25] revealed some statistically significant differences (A, top). Comparison of the set of upregulated genes with the complete set of Drosophila genes revealed some groups of genes that are highly enriched in this cohort (top). In addition, KEGG pathway analysis identified the oxidative phosphorylation as being significantly enriched in this cohort of genes. The cohort of downregulated genes includes two functional groups, being significantly different from the total number of genes (A, bottom). The corresponding lists are summarized in additional file 5. Venn diagram analyses showed that only a small minority of the up- or downregulated genes are classical immune relevant genes (additional file 1). Similarly, the intersection with those genes that are regulated upon an experimental infection with the insect pathogen Erwinia carotovora in two different epithelial tissues are rather small (B). In addition, small groups of genes are identical if compared with genes upregulated in the trachea following the same experimental manipulation (IMD-activation) and with susceptibility genes for gut infection (C), SG IMD up means the cohort of genes upregulated following IMD activation (see additional files 1, 2, 3, 4 and 5), gut and trachea Erwinia up means the groups of genes upregulated in the trachea or the gut following infection with Erwinia carotovora [9,27], trachea IMD up means the list of genes upregulated following IMD activation in the larval trachea [27], gut infection susceptibility means those genes that have been identified as having an influence on oral infection in a genome wide RNAi screen [8].
Figure 5
Verification of differential expression of some highly interesting genes, which are regulated following IMD-activation within the salivary glands. RNA is isolated from pure salivary glands (freed of contaminating material) of both experimental groups and the results are the means of at least three independent experiments performed in duplicate. All genes are transcribed at significantly higher rates in the experimental tissues compared with controls (p < 0.05).
Similar articles
- Tissue-autonomous immune response regulates stress signaling during hypertrophy.
Krautz R, Khalili D, Theopold U. Krautz R, et al. Elife. 2020 Dec 30;9:e64919. doi: 10.7554/eLife.64919. Elife. 2020. PMID: 33377870 Free PMC article. - Molecular architecture of the fruit fly's airway epithelial immune system.
Wagner C, Isermann K, Fehrenbach H, Roeder T. Wagner C, et al. BMC Genomics. 2008 Sep 29;9:446. doi: 10.1186/1471-2164-9-446. BMC Genomics. 2008. PMID: 18823557 Free PMC article. - Tsetse fly tolerance to T. brucei infection: transcriptome analysis of trypanosome-associated changes in the tsetse fly salivary gland.
Matetovici I, Caljon G, Van Den Abbeele J. Matetovici I, et al. BMC Genomics. 2016 Nov 25;17(1):971. doi: 10.1186/s12864-016-3283-0. BMC Genomics. 2016. PMID: 27884110 Free PMC article. - The Drosophila imd signaling pathway.
Myllymäki H, Valanne S, Rämet M. Myllymäki H, et al. J Immunol. 2014 Apr 15;192(8):3455-62. doi: 10.4049/jimmunol.1303309. J Immunol. 2014. PMID: 24706930 Review. - Drosophila immune response: From systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract.
Charroux B, Royet J. Charroux B, et al. Fly (Austin). 2010 Jan-Mar;4(1):40-7. doi: 10.4161/fly.4.1.10810. Epub 2010 Jan 2. Fly (Austin). 2010. PMID: 20383054 Review.
Cited by
- Impaired Wnt signaling in dopamine containing neurons is associated with pathogenesis in a rotenone triggered Drosophila Parkinson's disease model.
Stephano F, Nolte S, Hoffmann J, El-Kholy S, von Frieling J, Bruchhaus I, Fink C, Roeder T. Stephano F, et al. Sci Rep. 2018 Feb 5;8(1):2372. doi: 10.1038/s41598-018-20836-w. Sci Rep. 2018. PMID: 29403026 Free PMC article. - A critical role for the Drosophila dopamine D1-like receptor Dop1R2 at the onset of metamorphosis.
Regna K, Kurshan PT, Harwood BN, Jenkins AM, Lai CQ, Muskavitch MA, Kopin AS, Draper I. Regna K, et al. BMC Dev Biol. 2016 May 16;16(1):15. doi: 10.1186/s12861-016-0115-z. BMC Dev Biol. 2016. PMID: 27184815 Free PMC article. - Toll-8/Tollo negatively regulates antimicrobial response in the Drosophila respiratory epithelium.
Akhouayri I, Turc C, Royet J, Charroux B. Akhouayri I, et al. PLoS Pathog. 2011 Oct;7(10):e1002319. doi: 10.1371/journal.ppat.1002319. Epub 2011 Oct 13. PLoS Pathog. 2011. PMID: 22022271 Free PMC article. - Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila.
Hoffmann J, Romey R, Fink C, Yong L, Roeder T. Hoffmann J, et al. Aging (Albany NY). 2013 Apr;5(4):315-27. doi: 10.18632/aging.100553. Aging (Albany NY). 2013. PMID: 23765091 Free PMC article. - Prostaglandins and their receptors in insect biology.
Stanley D, Kim Y. Stanley D, et al. Front Endocrinol (Lausanne). 2011 Dec 30;2:105. doi: 10.3389/fendo.2011.00105. eCollection 2011. Front Endocrinol (Lausanne). 2011. PMID: 22654840 Free PMC article.
References
- Silverman N, Paquette N. Immunology. The right resident bugs. Science (New York, NY) 2008;319(5864):734–735. - PubMed
- Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. The EMBO journal. 1998;17(5):1217–1227. doi: 10.1093/emboj/17.5.1217. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases