A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62 - PubMed (original) (raw)
A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62
Alexandria Lau et al. Mol Cell Biol. 2010 Jul.
Abstract
In response to stress, cells can utilize several cellular processes, such as autophagy, which is a bulk-lysosomal degradation pathway, to mitigate damages and increase the chances of cell survival. Deregulation of autophagy causes upregulation of p62 and the formation of p62-containing aggregates, which are associated with neurodegenerative diseases and cancer. The Nrf2-Keap1 pathway functions as a critical regulator of the cell's defense mechanism against oxidative stress by controlling the expression of many cellular protective proteins. Under basal conditions, Nrf2 is ubiquitinated by the Keap1-Cul3-E3 ubiquitin ligase complex and targeted to the 26S proteasome for degradation. Upon induction, the activity of the E3 ubiquitin ligase is inhibited through the modification of cysteine residues in Keap1, resulting in the stabilization and activation of Nrf2. In this current study, we identified the direct interaction between p62 and Keap1 and the residues required for the interaction have been mapped to 349-DPSTGE-354 in p62 and three arginines in the Kelch domain of Keap1. Accumulation of endogenous p62 or ectopic expression of p62 sequesters Keap1 into aggregates, resulting in the inhibition of Keap1-mediated Nrf2 ubiquitination and its subsequent degradation by the proteasome. In contrast, overexpression of mutated p62, which loses its ability to interact with Keap1, had no effect on Nrf2 stability, demonstrating that p62-mediated Nrf2 upregulation is Keap1 dependent. These findings demonstrate that autophagy deficiency activates the Nrf2 pathway in a noncanonical cysteine-independent mechanism.
Figures
FIG. 1.
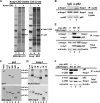
The interaction between p62 and Keap1 and the domains that are required for the interaction. (A) Identification of the Keap1-interacting protein p62. Two stable cell lines, MDA-MB-231 and Keap1−/−, that expressed either vector control (−) or CBD-tagged Keap1 (+) were used in a pulldown assay using chitin beads. Samples were run on an SDS-PAGE gel and silver stained. The distinct band was excised and identified as p62. L, protein ladder. (B) Interaction of endogenous p62 and Keap1. Cell lysates from HEK293 cells were immunoprecipitated (IP) using an antibody against p62 or IgG (negative control). The resulting immunoprecipitates were subjected to SDS-PAGE and analyzed by immunoblotting with a Keap1 antibody. (C) The Keap1-interacting domain of p62 is 349-DPSTGE-354. p62 deletion mutants (p62-380 [deletion of aa 381 to the C terminus] and p62-300 [aa 301 to the C terminus]), along with p62 wild-type (p62-WT) and a mutant in which amino acids 349 to 354 were changed to alanines (p62-M), were constructed into an expression vector containing a T7 promoter. In vitro transcription and translation were performed to generate 35S-labeled full-length p62-WT, p62-M, and its deletion proteins. Each of these proteins was incubated with GST-Keap1, which was produced and purified from E. coli bacteria. Keap1-associated proteins were pulled down using glutathione beads. Equivalent amounts of p62 proteins were used in the GST pulldown assay. (D) The Kelch domain of Keap1 is the binding domain of p62. Full-length Keap1, its deletion proteins (N-terminal [ΔN], BTB domain [ΔB], linker domain [ΔL], Kelch domain [ΔK], and the C terminus [ΔC]), and p62-WT were generated by in vitro transcription and translation methods. Each of the 35S-labeled Keap1 proteins were incubated with p62-WT followed by nickel affinity chromatography. Equal amounts of Keap1 proteins were used for this pulldown assay. (E) Immunoprecipitation analysis was performed using cell lysates of HEK293 cells cotransfected with an expression vector for either p62-WT or p62-M, along with an expression vector for Keap1. Expression of each protein was detected using anti-Keap1, anti-myc (for p62 proteins), and anti-β-actin (for loading control) antibodies. (F) Three arginines in the Kelch domain of Keap1 are responsible for the binding of p62. The three arginine residues at positions 380, 415, and 483 were replaced with alanine residues using site-directed mutagenesis. The indicated constructs were coexpressed with myc-p62 in HEK293 cells by transfection, and lysates were immunoprecipitated using an anti-myc antibody. Precipitates were subjected to SDS-PAGE and immunoblotted with the indicated antibodies.
FIG. 2.
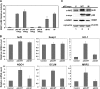
p62 upregulated the Nrf2 signaling pathway. (A) p62 regulates the transcriptional activity of Nrf2. Different amounts of the expression plasmids of p62-WT or p62-M were transfected into HEK293 cells along with the NQO1-ARE promoter firefly luciferase and R enilla luciferase as an internal control. As a positive control, cells were treated with known inducers of Nrf2, tBHQ, or SF overnight. Thirty-six hours posttransfection, both firefly and R enilla luciferase activities were measured. (B) Upregulation of Nrf2 downstream genes by p62. HEK293 cells were transfected with either the expression plasmid for vector, p62-WT, or p62-M. Forty-eight hours posttransfection, total mRNA was extracted using TRIzol, and qRT-PCR was performed to measure the mRNA levels of Nrf2, Keap1, HO-1, NQO1, GLCM, and MRP2. Values were normalized to GAPDH, and samples were done in triplicates. Data represented are the mean ± standard deviation (SD). (C) An increase in p62 caused an increase in Nrf2 protein levels. HEK293 cells were transfected with an expression vector for vector, p62-WT, or p62-M. Cell lysates were collected and subjected to immunoblot analysis using the following antibodies: anti-Nrf2, anti-myc (p62), anti-NQO1, and anti-β-actin.
FIG. 3.
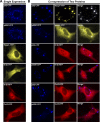
p62 sequestered Keap1 into aggregates. (A) The cellular localization of p62, Keap1, Nrf2, and Cul3. HEK293 cells were singly transfected with an expression vector for the fluorescently tagged protein. The subcellular localization of the proteins was monitored live. (B and C) The colocalization of proteins. Live imaging of the proteins was monitored with HEK293 cells that were cotransfected with expression vectors for the indicated fluorescently tagged proteins.
FIG. 3.
p62 sequestered Keap1 into aggregates. (A) The cellular localization of p62, Keap1, Nrf2, and Cul3. HEK293 cells were singly transfected with an expression vector for the fluorescently tagged protein. The subcellular localization of the proteins was monitored live. (B and C) The colocalization of proteins. Live imaging of the proteins was monitored with HEK293 cells that were cotransfected with expression vectors for the indicated fluorescently tagged proteins.
FIG. 4.
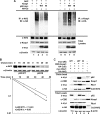
p62 decreased ubiquitination of Nrf2, leading to an increase in Nrf2 stability. (A) p62 decreased the ubiquitination of Nrf2 and increased the ubiquitination of Keap1. HEK293 cells were cotransfected with an expression vector for Nrf2, Keap1, HA-ubiquitin, and either p62-WT or p62-M. In vivo ubiquitination analysis was performed 48 h posttransfection. IB, immunoblotting. (B) p62 increased the half-life of Nrf2. HEK293 cells transfected with either p62-WT or p62-M were treated with 25 μM CHX for the indicated time periods (chase). Endogenous Nrf2 levels were detected by immunoblotting, and the intensity of the Nrf2 bands was quantified and plotted on a semilog graph. (C) p62-WT increased the association of Keap1, Cul3, and Rbx1. Immunoprecipitation analysis was conducted with HEK293 cells transfected with the indicated proteins. Nrf2 was included as a positive control.
FIG. 5.
Autophagy-defective cells sequestered Keap1 into aggregates. (A and B) Keap1 was sequestered into aggregates in primary autophagy-deficient cells. Atg5+/+, Atg5−/−, Beclin1+/+, and Beclin+/− iBMK cells stably expressing GFP alone or p62-GFP were transfected with CFP-tagged Keap1. Live imaging was taken 24 h posttransfection. (C and D) Endogenous Keap1 was sequestered into aggregates in primary autophagy-deficient cells. The cells were grown on coverslips and fixed in methanol. Indirect immunofluorescence staining was conducted using an antibody against GFP and Keap1.
Similar articles
- The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1.
Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. Komatsu M, et al. Nat Cell Biol. 2010 Mar;12(3):213-23. doi: 10.1038/ncb2021. Epub 2010 Feb 21. Nat Cell Biol. 2010. PMID: 20173742 - Keap1 degradation by autophagy for the maintenance of redox homeostasis.
Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Taguchi K, et al. Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13561-6. doi: 10.1073/pnas.1121572109. Epub 2012 Aug 7. Proc Natl Acad Sci U S A. 2012. PMID: 22872865 Free PMC article. - Tripartite motif 25 inhibits protein aggregate degradation during PRRSV infection by suppressing p62-mediated autophagy.
Ren J, Pei Q, Dong H, Wei X, Li L, Duan H, Zhang G, Zhang A. Ren J, et al. J Virol. 2024 Nov 19;98(11):e0143724. doi: 10.1128/jvi.01437-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480084 Free PMC article. - Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases.
Villeneuve NF, Lau A, Zhang DD. Villeneuve NF, et al. Antioxid Redox Signal. 2010 Dec 1;13(11):1699-712. doi: 10.1089/ars.2010.3211. Epub 2010 Aug 14. Antioxid Redox Signal. 2010. PMID: 20486766 Free PMC article. Review. - Novel target for treating Alzheimer's Diseases: Crosstalk between the Nrf2 pathway and autophagy.
Zhang W, Feng C, Jiang H. Zhang W, et al. Ageing Res Rev. 2021 Jan;65:101207. doi: 10.1016/j.arr.2020.101207. Epub 2020 Nov 1. Ageing Res Rev. 2021. PMID: 33144123 Review.
Cited by
- Innate immune evasion by filoviruses.
Basler CF. Basler CF. Virology. 2015 May;479-480:122-30. doi: 10.1016/j.virol.2015.03.030. Epub 2015 Apr 3. Virology. 2015. PMID: 25843618 Free PMC article. Review. - Reactive Oxygen Species and Nuclear Factor Erythroid 2-Related Factor 2 Activation in Diabetic Nephropathy: A Hidden Target.
Abdo S, Zhang SL, Chan JS. Abdo S, et al. J Diabetes Metab. 2015 May 10;6(6):10.4172/2155-6156.1000547. doi: 10.4172/2155-6156.1000547. J Diabetes Metab. 2015. PMID: 26213634 Free PMC article. - When autophagy meets cancer through p62/SQSTM1.
Puissant A, Fenouille N, Auberger P. Puissant A, et al. Am J Cancer Res. 2012;2(4):397-413. Epub 2012 Jun 28. Am J Cancer Res. 2012. PMID: 22860231 Free PMC article. - Cannabinoid-Induced Autophagy and Heme Oxygenase-1 Determine the Fate of Adipose Tissue-Derived Mesenchymal Stem Cells under Stressful Conditions.
Bublitz K, Böckmann S, Peters K, Hinz B. Bublitz K, et al. Cells. 2020 Oct 15;9(10):2298. doi: 10.3390/cells9102298. Cells. 2020. PMID: 33076330 Free PMC article. - Peroxisome proliferator-activated receptor δ agonist, HPP593, prevents renal necrosis under chronic ischemia.
Fedorova LV, Sodhi K, Gatto-Weis C, Puri N, Hinds TD Jr, Shapiro JI, Malhotra D. Fedorova LV, et al. PLoS One. 2013 May 15;8(5):e64436. doi: 10.1371/journal.pone.0064436. Print 2013. PLoS One. 2013. PMID: 23691217 Free PMC article.
References
- Ishii, T., K. Itoh, S. Takahashi, H. Sato, T. Yanagawa, Y. Katoh, S. Bannai, and M. Yamamoto. 2000. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275:16023-16029. - PubMed
- Kim, Y. C., H. Masutani, Y. Yamaguchi, K. Itoh, M. Yamamoto, and J. Yodoi. 2001. Hemin-induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J. Biol. Chem. 276:18399-18406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES006694/ES/NIEHS NIH HHS/United States
- R01ES015010/ES/NIEHS NIH HHS/United States
- ES006694/ES/NIEHS NIH HHS/United States
- RSG-07-154/PHS HHS/United States
- R01 ES015010/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases