Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea - PubMed (original) (raw)
. 2010 May 11;107(19):8818-23.
doi: 10.1073/pnas.0913533107. Epub 2010 Apr 26.
J R de la Torre, M G Klotz, H Urakawa, N Pinel, D J Arp, C Brochier-Armanet, P S G Chain, P P Chan, A Gollabgir, J Hemp, M Hügler, E A Karr, M Könneke, M Shin, T J Lawton, T Lowe, W Martens-Habbena, L A Sayavedra-Soto, D Lang, S M Sievert, A C Rosenzweig, G Manning, D A Stahl
Affiliations
- PMID: 20421470
- PMCID: PMC2889351
- DOI: 10.1073/pnas.0913533107
Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea
C B Walker et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Ammonia-oxidizing archaea are ubiquitous in marine and terrestrial environments and now thought to be significant contributors to carbon and nitrogen cycling. The isolation of Candidatus "Nitrosopumilus maritimus" strain SCM1 provided the opportunity for linking its chemolithotrophic physiology with a genomic inventory of the globally distributed archaea. Here we report the 1,645,259-bp closed genome of strain SCM1, revealing highly copper-dependent systems for ammonia oxidation and electron transport that are distinctly different from known ammonia-oxidizing bacteria. Consistent with in situ isotopic studies of marine archaea, the genome sequence indicates N. maritimus grows autotrophically using a variant of the 3-hydroxypropionate/4-hydroxybutryrate pathway for carbon assimilation, while maintaining limited capacity for assimilation of organic carbon. This unique instance of archaeal biosynthesis of the osmoprotectant ectoine and an unprecedented enrichment of multicopper oxidases, thioredoxin-like proteins, and transcriptional regulators points to an organism responsive to environmental cues and adapted to handling reactive copper and nitrogen species that likely derive from its distinctive biochemistry. The conservation of N. maritimus gene content and organization within marine metagenomes indicates that the unique physiology of these specialized oligophiles may play a significant role in the biogeochemical cycles of carbon and nitrogen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Proposed AOA respiratory pathway. Text indicates the described possible hydroxylamine (blue text and arrows) and nitroxyl (green) pathways. Red arrows indicate electron flow not involved in ammonia oxidation. Blue shading denotes blue copper-containing proteins. Pink box indicates possible alternative respiratory electron sink. Hexagons containing Q and QH2 represent the oxidized and reduced quinone pools, respectively.
Fig. 2.
Synteny plots comparing the N. maritimus genome with (A) the Cenarchaeum symbiosum A type genome, (B) crenarchaeal genome fragments, and (C) Sargasso Sea fosmids. Vertical gray bar indicates location of ribosomal RNA operon.
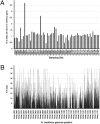
Fig. 3.
Comparison of N. maritimus genome to metagenomic sequence reads from GOS. (A) Percentage of reads from each GOS site that align to the N. maritimus genome by protein sequence similarity. (B) Number of GOS reads homologous to each N. maritimus protein-coding gene. Counts of 40 are mostly due to highly conserved proteins that include contaminants from other clades.
Similar articles
- Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. Hallam SJ, et al. PLoS Biol. 2006 Apr;4(4):e95. doi: 10.1371/journal.pbio.0040095. Epub 2006 Mar 21. PLoS Biol. 2006. PMID: 16533068 Free PMC article. - Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation.
Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, Schada von Borzyskowski L, Erb TJ, Stahl DA, Berg IA. Könneke M, et al. Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8239-44. doi: 10.1073/pnas.1402028111. Epub 2014 May 19. Proc Natl Acad Sci U S A. 2014. PMID: 24843170 Free PMC article. - Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume.
Baker BJ, Lesniewski RA, Dick GJ. Baker BJ, et al. ISME J. 2012 Dec;6(12):2269-79. doi: 10.1038/ismej.2012.64. Epub 2012 Jun 14. ISME J. 2012. PMID: 22695863 Free PMC article. - Nitrogen metabolism and kinetics of ammonia-oxidizing archaea.
Martens-Habbena W, Stahl DA. Martens-Habbena W, et al. Methods Enzymol. 2011;496:465-87. doi: 10.1016/B978-0-12-386489-5.00019-1. Methods Enzymol. 2011. PMID: 21514476 Review. - Ammonia-oxidizing archaea involved in nitrogen removal.
You J, Das A, Dolan EM, Hu Z. You J, et al. Water Res. 2009 Apr;43(7):1801-9. doi: 10.1016/j.watres.2009.01.016. Epub 2009 Jan 31. Water Res. 2009. PMID: 19232671 Review.
Cited by
- Effects of Different Land Use Types on Active Autotrophic Ammonia and Nitrite Oxidizers in Cinnamon Soils.
Wang X, Wang Y, Zhu F, Zhang C, Wang P, Zhang X. Wang X, et al. Appl Environ Microbiol. 2021 May 26;87(12):e0009221. doi: 10.1128/AEM.00092-21. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33837020 Free PMC article. - Microbial communities involved in biological ammonium removal from coal combustion wastewaters.
Vishnivetskaya TA, Fisher LS, Brodie GA, Phelps TJ. Vishnivetskaya TA, et al. Microb Ecol. 2013 Jul;66(1):49-59. doi: 10.1007/s00248-012-0152-5. Epub 2013 Jan 13. Microb Ecol. 2013. PMID: 23314095 - Biogeochemical and Microbial Variation across 5500 km of Antarctic Surface Sediment Implicates Organic Matter as a Driver of Benthic Community Structure.
Learman DR, Henson MW, Thrash JC, Temperton B, Brannock PM, Santos SR, Mahon AR, Halanych KM. Learman DR, et al. Front Microbiol. 2016 Mar 23;7:284. doi: 10.3389/fmicb.2016.00284. eCollection 2016. Front Microbiol. 2016. PMID: 27047451 Free PMC article. - NmPin from the marine thaumarchaeote Nitrosopumilus maritimus is an active membrane associated prolyl isomerase.
Hoppstock L, Trusch F, Lederer C, van West P, Koenneke M, Bayer P. Hoppstock L, et al. BMC Biol. 2016 Jun 27;14:53. doi: 10.1186/s12915-016-0274-1. BMC Biol. 2016. PMID: 27349962 Free PMC article. - The molecular basis of K+ exclusion by the Escherichia coli ammonium channel AmtB.
Hall JA, Yan D. Hall JA, et al. J Biol Chem. 2013 May 17;288(20):14080-14086. doi: 10.1074/jbc.M113.457952. Epub 2013 Apr 1. J Biol Chem. 2013. PMID: 23546877 Free PMC article.
References
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. - PubMed
- Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. - PubMed
- Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. - PubMed
- Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases