VLDL hydrolysis by LPL activates PPAR-alpha through generation of unbound fatty acids - PubMed (original) (raw)
VLDL hydrolysis by LPL activates PPAR-alpha through generation of unbound fatty acids
Maxwell A Ruby et al. J Lipid Res. 2010 Aug.
Abstract
Recent evidence suggests that lipoproteins serve as circulating reservoirs of peroxisomal proliferator activated receptor (PPAR) ligands that are accessible through lipolysis. The present study was conducted to determine the biochemical basis of PPAR-alpha activation by lipolysis products and their contribution to PPAR-alpha function in vivo. PPAR-alpha activation was measured in bovine aortic endothelial cells following treatment with human plasma, VLDL lipolysis products, or oleic acid. While plasma failed to activate PPAR-alpha, oleic acid performed similarly to VLDL lipolysis products. Therefore, fatty acids are likely to be the PPAR-alpha ligands generated by VLDL lipolysis. Indeed, unbound fatty acid concentration determined PPAR-alpha activation regardless of fatty acid source, with PPAR-alpha activation occurring only at unbound fatty acid concentrations that are unachievable under physiological conditions without lipase action. In mice, a synthetic lipase inhibitor (poloxamer-407) attenuated fasting-induced changes in expression of PPAR-alpha target genes. Apolipoprotein CIII (apoCIII), an endogenous inhibitor of lipoprotein and hepatic lipase, regulated access to the lipoprotein pool of PPAR-alpha ligands, because addition of exogenous apoCIII inhibited, and removal of endogenous apoCIII potentiated, lipolytic PPAR-alpha activation. These data suggest that the PPAR-alpha response is generated by unbound fatty acids released locally by lipase activity and not by circulating plasma fatty acids.
Figures
Fig. 1.
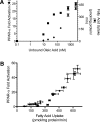
VLDL-derived fatty acids serve as potent PPAR-α ligands due to efficient delivery. BAEC were transfected with the PPAR α-LBD-GAL4 reporter system, as described in “Methods,” treated for 18 h, and cell lysate assayed for luciferase and β-galoctosidase activity. A: BAEC were exposed to various concentrations of LPL (1, 3, or 10 units/ml) and VLDL (1, 3, 10, or 30 μg/ml), oleic acid (0, 5, 10, or 20 μM at an unbound oleic acid concentration of 2450 nM), or plasma (0–5% v/v). This produced a range of NEFA concentrations, as measured in the cell culture media at the end of treatment. For oleic acid, a linear relationship exists between oleic acid added and NEFA concentration in the cell culture media (data not shown), such that ∼60% of NEFA added remains in the media following incubation with cells. PPAR-α activity is presented as percentage of activation by 10 µM Wy14643, a synthetic PPAR-α ligand. B: Transfected BAEC were incubated with VLDL (10 µg protein/ml) and LPL (10 units/ml) and increasing concentrations of albumin in triplicate. PPAR-α activity is expressed as fold activation over control.
Fig. 2.
Fatty acid uptake determines PPAR-α activation. A: In parallel experiments, BAECs were treated with oleic acid (90 μM) and varying concentrations of albumin, and PPAR-α activation and fatty acid uptake were determined as described in Methods. B: Varying total (0–180 μM) and unbound oleic acid concentrations (0–2,450 nM) were used to generate a range of fatty acid uptake. Fatty acid uptake above 300 pmol/mg protein/min displayed a strong linear relationship with PPAR-α activation (_r_2 = 0.98; P < 0.05).
Fig. 3.
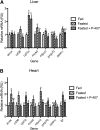
P-407 inhibits the transcriptional response to fasting in vivo. Following an initial 2 h fast, 9-week-old male C57Bl6 mice were treated with saline or P-407 (500 mg/kg, i.p.) and fasted for an additional 24 h. A group of saline-injected animals was fed ad libitum for the 24 h period. Abundance of mRNA for PPAR-α target genes was determined by RT-PCR and normalized to a control gene (gusb) in the liver (A) and heart (B). Groups not sharing a common superscript letter are significantly different (P < 0.05).
Fig. 4.
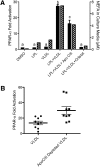
ApoCIII regulates access to lipoprotein-derived PPAR-α ligands. A: PPAR-α activation (solid bars) in BAEC were transfected as described in Fig. 1 and treated with LPL (10 units/ml), VLDL (10 µg protein/ml), Orlistat (10 µM), or apoCIII (3 μg/ml) as indicated for 18 h. Cell culture media was assayed for fatty acids. Groups not sharing a common superscript letter are significantly different (P < 0.05). B: BAEC were transfected as described in Fig. 1 and treated with total VLDL or apoCIII-depleted VLDL (10 μg protein/ml) and LPL (3 units/ml). The difference between the VLDL treatments was significant (P = 0.002) by paired two-tailed _t_-test.
Similar articles
- Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: evidence for an antiinflammatory role for lipoprotein lipase.
Ziouzenkova O, Perrey S, Asatryan L, Hwang J, MacNaul KL, Moller DE, Rader DJ, Sevanian A, Zechner R, Hoefler G, Plutzky J. Ziouzenkova O, et al. Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2730-5. doi: 10.1073/pnas.0538015100. Epub 2003 Feb 26. Proc Natl Acad Sci U S A. 2003. PMID: 12606719 Free PMC article. - Influence of lipoprotein lipase and hepatic lipase on the transformation of VLDL and HDL during lipolysis of VLDL.
Murdoch SJ, Breckenridge WC. Murdoch SJ, et al. Atherosclerosis. 1995 Dec;118(2):193-212. doi: 10.1016/0021-9150(95)05606-8. Atherosclerosis. 1995. PMID: 8770314 - Lipoprotein lipase liberates free fatty acids to inhibit HCV infection and prevent hepatic lipid accumulation.
Sun HY, Lin CC, Tsai PJ, Tsai WJ, Lee JC, Tsao CW, Cheng PN, Wu IC, Chiu YC, Chang TT, Young KC. Sun HY, et al. Cell Microbiol. 2017 Apr;19(4). doi: 10.1111/cmi.12673. Epub 2016 Nov 25. Cell Microbiol. 2017. PMID: 27665576 - [THE LIPOLYSIS IN PHYLOGENETICALLY EARLY LIPOPROTEINS OF LOW DENSITY AND MORE LATER LIPOPROTEINS OF VERY LOW DENSITY: FUNCTION AND DIAGNOSTIC VALUE OF APOE AND APOC-III].
Rozhkova TA, Titov VN, Amelyushkina VA, Kaba SI, Kukhartchuk VV. Rozhkova TA, et al. Klin Lab Diagn. 2015 Dec;60(12):4-14. Klin Lab Diagn. 2015. PMID: 27032246 Review. Russian. - Regulation of energy metabolism by long-chain fatty acids.
Nakamura MT, Yudell BE, Loor JJ. Nakamura MT, et al. Prog Lipid Res. 2014 Jan;53:124-44. doi: 10.1016/j.plipres.2013.12.001. Epub 2013 Dec 18. Prog Lipid Res. 2014. PMID: 24362249 Review.
Cited by
- Early Life Exposure to Fructose Alters Maternal, Fetal and Neonatal Hepatic Gene Expression and Leads to Sex-Dependent Changes in Lipid Metabolism in Rat Offspring.
Clayton ZE, Vickers MH, Bernal A, Yap C, Sloboda DM. Clayton ZE, et al. PLoS One. 2015 Nov 12;10(11):e0141962. doi: 10.1371/journal.pone.0141962. eCollection 2015. PLoS One. 2015. PMID: 26562417 Free PMC article. - Aberrant LPL Expression, Driven by STAT3, Mediates Free Fatty Acid Metabolism in CLL Cells.
Rozovski U, Grgurevic S, Bueso-Ramos C, Harris DM, Li P, Liu Z, Wu JY, Jain P, Wierda W, Burger J, O'Brien S, Jain N, Ferrajoli A, Keating MJ, Estrov Z. Rozovski U, et al. Mol Cancer Res. 2015 May;13(5):944-53. doi: 10.1158/1541-7786.MCR-14-0412. Epub 2015 Mar 2. Mol Cancer Res. 2015. PMID: 25733697 Free PMC article. - Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4α and PPARα.
Bushkofsky JR, Maguire M, Larsen MC, Fong YH, Jefcoate CR. Bushkofsky JR, et al. Arch Biochem Biophys. 2016 May 1;597:30-47. doi: 10.1016/j.abb.2016.03.030. Epub 2016 Mar 29. Arch Biochem Biophys. 2016. PMID: 27036855 Free PMC article. - Functional Role of PPARs in Ruminants: Potential Targets for Fine-Tuning Metabolism during Growth and Lactation.
Bionaz M, Chen S, Khan MJ, Loor JJ. Bionaz M, et al. PPAR Res. 2013;2013:684159. doi: 10.1155/2013/684159. Epub 2013 Apr 29. PPAR Res. 2013. PMID: 23737762 Free PMC article. - Anti-Hypertensive Action of Fenofibrate via UCP2 Upregulation Mediated by PPAR Activation in Baroreflex Afferent Pathway.
Guan J, Zhao M, He C, Li X, Li Y, Sun J, Wang W, Cui YL, Zhang Q, Li BY, Qiao GF. Guan J, et al. Neurosci Bull. 2019 Feb;35(1):15-24. doi: 10.1007/s12264-018-0271-1. Epub 2018 Sep 1. Neurosci Bull. 2019. PMID: 30173356 Free PMC article.
References
- Brown J. D., Plutzky J. 2007. Peroxisome proliferator-activated receptors as transcriptional nodal points and therapeutic targets. Circulation. 115: 518–533. - PubMed
- Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., et al. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA. 94: 4318–4323. - PMC - PubMed
- Lee C. H., Kang K., Mehl I. R., Nofsinger R., Alaynick W. A., Chong L. W., Rosenfeld J. M., Evans R. M. 2006. Peroxisome proliferator-activated receptor delta promotes very low-density lipoprotein-derived fatty acid catabolism in the macrophage. Proc. Natl. Acad. Sci. USA. 103: 2434–2439. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL048743/HL/NHLBI NIH HHS/United States
- R01 HL071745/HL/NHLBI NIH HHS/United States
- P0-1HL48743/HL/NHLBI NIH HHS/United States
- R0-1HL071745/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources