The peopling of Europe from the mitochondrial haplogroup U5 perspective - PubMed (original) (raw)
The peopling of Europe from the mitochondrial haplogroup U5 perspective
Boris Malyarchuk et al. PLoS One. 2010.
Abstract
It is generally accepted that the most ancient European mitochondrial haplogroup, U5, has evolved essentially in Europe. To resolve the phylogeny of this haplogroup, we completely sequenced 113 mitochondrial genomes (79 U5a and 34 U5b) of central and eastern Europeans (Czechs, Slovaks, Poles, Russians and Belorussians), and reconstructed a detailed phylogenetic tree, that incorporates previously published data. Molecular dating suggests that the coalescence time estimate for the U5 is approximately 25-30 thousand years (ky), and approximately 16-20 and approximately 20-24 ky for its subhaplogroups U5a and U5b, respectively. Phylogeographic analysis reveals that expansions of U5 subclusters started earlier in central and southern Europe, than in eastern Europe. In addition, during the Last Glacial Maximum central Europe (probably, the Carpathian Basin) apparently represented the area of intermingling between human flows from refugial zones in the Balkans, the Mediterranean coastline and the Pyrenees. Age estimations amounting for many U5 subclusters in eastern Europeans to approximately 15 ky ago and less are consistent with the view that during the Ice Age eastern Europe was an inhospitable place for modern humans.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
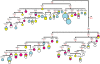
Figure 1. Complete mtDNA phylogenetic tree of haplogroup U5.
This schematic tree is based on two phylogenetic trees (for U5a and U5b) presented in Figures S1 and S2, respectively. Nomenclature of hg U5 subclusters was applied in accordance with the classification described in the Table S1. Some unnamed tips are singular haplotypes (e.g. U5a1*) aggregated for the purposes of illustration. Time estimates (in ky) shown for mtDNA subclusters are based on the complete mitochondrial genome clock . The size of each circle is proportional to the number of individuals sharing the corresponding haplotype, with the smallest size corresponding to one individual. Geographical origins are indicated by different colors: eastern European – in blue, central European – in yellow, Mediterranean and western European – in fuchsia, and others (i.e. of unknown population origin) – in white. More detailed information on the origin of population samples can be found in Figures S1 and S2.
Similar articles
- Mitogenomes from two uncommon haplogroups mark late glacial/postglacial expansions from the near east and neolithic dispersals within Europe.
Olivieri A, Pala M, Gandini F, Hooshiar Kashani B, Perego UA, Woodward SR, Grugni V, Battaglia V, Semino O, Achilli A, Richards MB, Torroni A. Olivieri A, et al. PLoS One. 2013 Jul 31;8(7):e70492. doi: 10.1371/journal.pone.0070492. Print 2013. PLoS One. 2013. PMID: 23936216 Free PMC article. - Mitochondrial DNA phylogeny in Eastern and Western Slavs.
Malyarchuk B, Grzybowski T, Derenko M, Perkova M, Vanecek T, Lazur J, Gomolcak P, Tsybovsky I. Malyarchuk B, et al. Mol Biol Evol. 2008 Aug;25(8):1651-8. doi: 10.1093/molbev/msn114. Epub 2008 May 13. Mol Biol Evol. 2008. PMID: 18477584 - The history of Slavs inferred from complete mitochondrial genome sequences.
Mielnik-Sikorska M, Daca P, Malyarchuk B, Derenko M, Skonieczna K, Perkova M, Dobosz T, Grzybowski T. Mielnik-Sikorska M, et al. PLoS One. 2013;8(1):e54360. doi: 10.1371/journal.pone.0054360. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23342138 Free PMC article. - The archaeogenetics of Europe.
Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB. Soares P, et al. Curr Biol. 2010 Feb 23;20(4):R174-83. doi: 10.1016/j.cub.2009.11.054. Curr Biol. 2010. PMID: 20178764 Review. - Ancestry of modern Europeans: contributions of ancient DNA.
Lacan M, Keyser C, Crubézy E, Ludes B. Lacan M, et al. Cell Mol Life Sci. 2013 Jul;70(14):2473-87. doi: 10.1007/s00018-012-1180-5. Epub 2012 Oct 11. Cell Mol Life Sci. 2013. PMID: 23052219 Free PMC article. Review.
Cited by
- Evolution and dispersal of mitochondrial DNA haplogroup U5 in Northern Europe: insights from an unsupervised learning approach to phylogeography.
Kristjansson D, Bohlin J, Nguyen TT, Jugessur A, Schurr TG. Kristjansson D, et al. BMC Genomics. 2022 May 7;23(1):354. doi: 10.1186/s12864-022-08572-y. BMC Genomics. 2022. PMID: 35525961 Free PMC article. - Ancient DNA reveals prehistoric gene-flow from siberia in the complex human population history of North East Europe.
Der Sarkissian C, Balanovsky O, Brandt G, Khartanovich V, Buzhilova A, Koshel S, Zaporozhchenko V, Gronenborn D, Moiseyev V, Kolpakov E, Shumkin V, Alt KW, Balanovska E, Cooper A, Haak W; Genographic Consortium. Der Sarkissian C, et al. PLoS Genet. 2013;9(2):e1003296. doi: 10.1371/journal.pgen.1003296. Epub 2013 Feb 14. PLoS Genet. 2013. PMID: 23459685 Free PMC article. - Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia.
Derenko M, Malyarchuk B, Grzybowski T, Denisova G, Rogalla U, Perkova M, Dambueva I, Zakharov I. Derenko M, et al. PLoS One. 2010 Dec 21;5(12):e15214. doi: 10.1371/journal.pone.0015214. PLoS One. 2010. PMID: 21203537 Free PMC article. - The paradox of HBV evolution as revealed from a 16th century mummy.
Patterson Ross Z, Klunk J, Fornaciari G, Giuffra V, Duchêne S, Duggan AT, Poinar D, Douglas MW, Eden JS, Holmes EC, Poinar HN. Patterson Ross Z, et al. PLoS Pathog. 2018 Jan 4;14(1):e1006750. doi: 10.1371/journal.ppat.1006750. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29300782 Free PMC article. - Ancient mitochondrial lineages support the prehistoric maternal root of Basques in Northern Iberian Peninsula.
Palencia-Madrid L, Cardoso S, Keyser C, López-Quintana JC, Guenaga-Lizasu A, de Pancorbo MM. Palencia-Madrid L, et al. Eur J Hum Genet. 2017 May;25(5):631-636. doi: 10.1038/ejhg.2017.24. Epub 2017 Mar 8. Eur J Hum Genet. 2017. PMID: 28272540 Free PMC article.
References
- Richards MB, Macaulay VA, Bandelt H-J, Sykes BC. Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet. 1998;62:241–260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources