Assembly, structure, and function of the 26S proteasome - PubMed (original) (raw)
Review
Assembly, structure, and function of the 26S proteasome
Lynn Bedford et al. Trends Cell Biol. 2010 Jul.
Abstract
The 26S proteasome is a large multiprotein complex involved in the regulated degradation of ubiquitinated proteins in the cell. The 26S proteasome has been shown to control an increasing number of essential biochemical mechanisms of the cellular lifecycle including DNA synthesis, repair, transcription, translation, and cell signal transduction. Concurrently, it is increasingly seen that malfunction of the ubiquitin proteasome system contributes to the pathogenesis of disease. The recent identification of four molecular chaperones, in addition to five previously identified chaperones, have provided mechanistic insight into how this cellular megastructure is assembled in the cell. These data, together with new insights into the structure and function of the proteasome, provide a much better understanding of this complex protease.
Crown Copyright 2010. Published by Elsevier Ltd. All rights reserved.
Figures
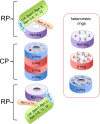
Figure 1
Composition of the 26S proteasome. Proteasome is formed by two regulatory particles abutting cylindrically shaped core particle. Core particle is formed by two α-rings and two β-rings. Regulatory particle consists of a base complex and a lid complex. Rpn10 is at the interface between these complexes (shown in light blue). The lid (green) contains contains indicated subunits and its function is not well understood, with the exception of rpn11, which functions as a de-ubiquitinating enzyme. The base contains a ring formed by six AAA-ATPases, Rpt1–6 (purple ring), and the subunits Rpn1, Rpn2 and Rpn13 (shown as orange box; see text for more details). The proteasome contains three heteromeric ring structures, each present twice. The α and β-ring are formed by seven subunits; scissors indicate the active sites and the dots in the a ring show the binding pockets for the Rpt tails (see text). The pocket between α7 and α1 lacks a Lys typically found in the pocket and might not harbour an Rpt C-terminus. The Rpt-ring is formed by six AAA-ATPases.
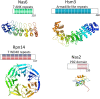
Figure 2
(A) Domain topology of Nas6, Rpn14, Hsm3 and Nas2. Below the topology is shown a 3D-structure of the domains to illustrate the structural difference. Only for Nas6 a structure been determined, for the others the structure of a similar domain from a different protein have been used (pdb coordinates used 1IXV for Nas6, 2H12 for WD40 repeat, 3GRL for Armadillo like repeats, 1G9O for PDZ domain). Hsm3 shows little conservation and detailed analysis of the domain can be found in Le Tallec et al.. Surprisingly, while structurally unrelated, these chaperones bind to the same small C-domain in the AAA-ATPases. (B) The domain topology of the proteasomal AAA-ATPases. cc is coiled coil region, OB is the OB-domain, ATPase is the ATPase domain containing the walker A and walker B motifs and the C indicates the C-domain, typically found in AAA-ATPases behind the ATPase domain. The structure shown is a ring formed by the ATPase (Blue) and C-domains (red) of six proteasome-activating nucleotidase (PAN) AAA-ATPases from the archaea Methanocaldococcus jannaschii. The CP cartoon is shown to illustrate the expected interface of the CP and the ATPase ring. Cartoons and structures are not drawn to scale.
Figure 2
(A) Domain topology of Nas6, Rpn14, Hsm3 and Nas2. Below the topology is shown a 3D-structure of the domains to illustrate the structural difference. Only for Nas6 a structure been determined, for the others the structure of a similar domain from a different protein have been used (pdb coordinates used 1IXV for Nas6, 2H12 for WD40 repeat, 3GRL for Armadillo like repeats, 1G9O for PDZ domain). Hsm3 shows little conservation and detailed analysis of the domain can be found in Le Tallec et al.. Surprisingly, while structurally unrelated, these chaperones bind to the same small C-domain in the AAA-ATPases. (B) The domain topology of the proteasomal AAA-ATPases. cc is coiled coil region, OB is the OB-domain, ATPase is the ATPase domain containing the walker A and walker B motifs and the C indicates the C-domain, typically found in AAA-ATPases behind the ATPase domain. The structure shown is a ring formed by the ATPase (Blue) and C-domains (red) of six proteasome-activating nucleotidase (PAN) AAA-ATPases from the archaea Methanocaldococcus jannaschii. The CP cartoon is shown to illustrate the expected interface of the CP and the ATPase ring. Cartoons and structures are not drawn to scale.
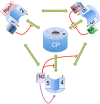
Figure 3
Cartoon showing the components known to be involved in the RP-assembly. The pathways and order of events are still unclear. Therefore an interaction map is displayed instead. The known potential interactions between the different complexes are shown in green. The chaperones are known to interfere with the interactions indicated in red, suggesting these proteins have a quality control role in assembly. Nas2 has also been shown to stabilize Rpt4 and 5 (purple arrow). In the middle of the cartoon are the seven pockets of the CP surrounding the gate (which is shown in an open conformation, although generally this is closed without activators bound to the CP). The dark pocket indicates the only pocket without a conserved positive charge expected not to be able to host a tail. Rpt tails that dock in the pockets are shown as dark orange extensions from the Rpt proteins. Numbers 1 to 6 indicate the different Rpt proteins, H3 Hsm3, R14 rpn14, N6 Nas6 and N2 Nas2.
Similar articles
- The recognition of ubiquitinated proteins by the proteasome.
Grice GL, Nathan JA. Grice GL, et al. Cell Mol Life Sci. 2016 Sep;73(18):3497-506. doi: 10.1007/s00018-016-2255-5. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137187 Free PMC article. Review. - The complexity of recognition of ubiquitinated substrates by the 26S proteasome.
Ciechanover A, Stanhill A. Ciechanover A, et al. Biochim Biophys Acta. 2014 Jan;1843(1):86-96. doi: 10.1016/j.bbamcr.2013.07.007. Epub 2013 Jul 18. Biochim Biophys Acta. 2014. PMID: 23872423 Review. - The mechanism for molecular assembly of the proteasome.
Sahara K, Kogleck L, Yashiroda H, Murata S. Sahara K, et al. Adv Biol Regul. 2014 Jan;54:51-8. doi: 10.1016/j.jbior.2013.09.010. Epub 2013 Oct 8. Adv Biol Regul. 2014. PMID: 24145026 Review. - Structure, assembly and homeostatic regulation of the 26S proteasome.
Xie Y. Xie Y. J Mol Cell Biol. 2010 Dec;2(6):308-17. doi: 10.1093/jmcb/mjq030. Epub 2010 Oct 7. J Mol Cell Biol. 2010. PMID: 20930034 Review. - Ubiquitin recognition by the proteasome.
Saeki Y. Saeki Y. J Biochem. 2017 Feb 1;161(2):113-124. doi: 10.1093/jb/mvw091. J Biochem. 2017. PMID: 28069863 Review.
Cited by
- Comprehensive analysis of bulk and single-cell RNA sequencing data reveals Schlafen-5 (SLFN5) as a novel prognosis and immunity.
Wu YJ, Chiao CC, Chuang PK, Hsieh CB, Ko CY, Ko CC, Chang CF, Chen TY, Nguyen NUN, Hsu CC, Chu TH, Fang CC, Tsai HY, Tsai HC, Anuraga G, Ta HDK, Xuan DTM, Kumar S, Dey S, Wulandari FS, Manalu RT, Ly NP, Wang CY, Lee YK. Wu YJ, et al. Int J Med Sci. 2024 Sep 9;21(12):2348-2364. doi: 10.7150/ijms.97975. eCollection 2024. Int J Med Sci. 2024. PMID: 39310264 Free PMC article. - Prion degradation pathways: Potential for therapeutic intervention.
Goold R, McKinnon C, Tabrizi SJ. Goold R, et al. Mol Cell Neurosci. 2015 May;66(Pt A):12-20. doi: 10.1016/j.mcn.2014.12.009. Epub 2015 Jan 10. Mol Cell Neurosci. 2015. PMID: 25584786 Free PMC article. Review. - Native Gel Approaches in Studying Proteasome Assembly and Chaperones.
Roelofs J, Suppahia A, Waite KA, Park S. Roelofs J, et al. Methods Mol Biol. 2018;1844:237-260. doi: 10.1007/978-1-4939-8706-1_16. Methods Mol Biol. 2018. PMID: 30242714 Free PMC article. - Stable incorporation of ATPase subunits into 19 S regulatory particle of human proteasome requires nucleotide binding and C-terminal tails.
Lee SH, Moon JH, Yoon SK, Yoon JB. Lee SH, et al. J Biol Chem. 2012 Mar 16;287(12):9269-79. doi: 10.1074/jbc.M111.316208. Epub 2012 Jan 24. J Biol Chem. 2012. PMID: 22275368 Free PMC article. - The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation.
Kumar B, Kim YC, DeMartino GN. Kumar B, et al. J Biol Chem. 2010 Dec 10;285(50):39523-35. doi: 10.1074/jbc.M110.153627. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937828 Free PMC article.
References
- Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
- Murata S, et al. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. - PubMed
- Marques AJ, et al. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F-0702/PUK_/Parkinson's UK/United Kingdom
- G-4055/PUK_/Parkinson's UK/United Kingdom
- P20 RR016475/RR/NCRR NIH HHS/United States
- P20 RR016475-09/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources