The "O" class: crafting clinical care with FoxO transcription factors - PubMed (original) (raw)
Review
The "O" class: crafting clinical care with FoxO transcription factors
Kenneth Maiese et al. Adv Exp Med Biol. 2009.
Abstract
Forkhead Transcription Factors: Vital Elements in Biology and Medicine provides a unique platform for the presentation of novel work and new insights into the vital role that forkhead transcription factors play in both cellular physiology as well as clinical medicine. Internationally recognized investigators provide their insights and perspectives for a number of forkhead genes and proteins that may have the greatest impact for the development of new strategies for a broad array of disorders that can involve aging, cancer, cardiac function, neurovascular integrity, fertility, stem cell differentiation, cellular metabolism, and immune system regulation. Yet, the work clearly sets a precedent for the necessity to understand the cellular and molecular function of forkhead proteins since this family of transcription factors can limit as well as foster disease progression depending upon the cellular environment. With this in mind, our concluding chapter for Forkhead Transcription Factors: Vital Elements in Biology andMedicine offers to highlight both the diversity and complexity of the forkhead transcription family by focusing upon the mammalian forkhead transcription factors of the O class (FoxOs) that include FoxO1, FoxO3, FoxO4, and FoxO6. FoxO proteins are increasingly considered to represent unique cellular targets that can control numerous processes such as angiogenesis, cardiovascular development, vascular tone, oxidative stress, stem cell proliferation, fertility, and immune surveillance. Furthermore, FoxO transcription factors are exciting considerations for disorders such as cancer in light of their pro-apoptotic and inhibitory cell cycle effects as well as diabetes mellitus given the close association FoxOs hold with cellular metabolism. In addition, these transcription factors are closely integrated with several novel signal transduction pathways, such as erythropoietin and Wnt proteins, that may influence the ability of FoxOs to lead to cell survival or cell injury. Further understanding of both the function and intricate nature of the forkhead transcription factor family, and in particular the FoxO proteins, should allow selective regulation of cellular development or cellular demise for the generation of successful future clinical strategies and patient well-being.
Figures
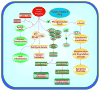
Figure 1
Posttranslational modulation of FoxO proteins is associated with intricate cellular signal transduction pathways. Posttranslational modulation of FoxO proteins involves pathways associated with phosphorylation, acetylation, and ubiquitylation. Protein kinase B (Akt) can prevent cellular apoptosis through the phosphorylation of FoxO proteins and phosphorylation (p) of FoxO proteins will inhibit FoxO transcription factors through cytoplasmic localization by association with 14-3-3 proteins and prevent the transcription of target genes that lead to apoptosis. If activated, FoxOs can prevent inflammatory cell activation through the inhibition of nuclear factor-κB (NF-κB) and may control inflammatory cell activation through membrane phosphatidylserine (PS) externalization. FoxO proteins can lead to apoptotic death pathways that involve mitochondrial (Mito) release of cytochrome c (Cyto c) and caspase activation through a Fas-mediated ligand (Fas L) death pathway, tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL), BH3-only proteins Noxa and Bim, or p53. Cell cycle inhibition that blocks tumor growth through FoxO protein activation may require c-myc, p27, and NF-κB.
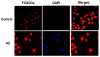
Figure 2
During amyloid (Aβ1-42) exposure in inflammatory microglial cells, FoxO3a translocates to the cell nucleus to govern an initial activation and proliferation of microglial cells. Microglia were followed at 6 hours after Aβ1-42 (10 μM) (Aβ) administration with immunofluorescent staining for FoxO3a (Texas-red). Nuclei of microglia were counterstained with DAPI. In merged images, control cells have readily visible nuclei (white in color) that illustrate absence of FoxO3a in the nucleus. In contrast, merged images after Aβ1-42 (10 μM) exposure are not visible (red in color) and demonstrate translocation of FoxO3a to the nucleus. Control = untreated microglia.
Similar articles
- A fork in the path: Developing therapeutic inroads with FoxO proteins.
Maiese K, Hou J, Chong ZZ, Shang YC. Maiese K, et al. Oxid Med Cell Longev. 2009 Jul-Aug;2(3):119-29. doi: 10.4161/oxim.2.3.8916. Oxid Med Cell Longev. 2009. PMID: 20592766 Free PMC article. Review. - A "FOXO" in sight: targeting Foxo proteins from conception to cancer.
Maiese K, Chong ZZ, Shang YC, Hou J. Maiese K, et al. Med Res Rev. 2009 May;29(3):395-418. doi: 10.1002/med.20139. Med Res Rev. 2009. PMID: 18985696 Free PMC article. Review. - OutFOXOing disease and disability: the therapeutic potential of targeting FoxO proteins.
Maiese K, Chong ZZ, Shang YC. Maiese K, et al. Trends Mol Med. 2008 May;14(5):219-27. doi: 10.1016/j.molmed.2008.03.002. Epub 2008 Apr 9. Trends Mol Med. 2008. PMID: 18403263 Free PMC article. Review. - Oxidative stress: Biomarkers and novel therapeutic pathways.
Maiese K, Chong ZZ, Hou J, Shang YC. Maiese K, et al. Exp Gerontol. 2010 Mar;45(3):217-34. doi: 10.1016/j.exger.2010.01.004. Epub 2010 Jan 11. Exp Gerontol. 2010. PMID: 20064603 Free PMC article. Review. - Should I stay or should I go: beta-catenin decides under stress.
Hoogeboom D, Burgering BM. Hoogeboom D, et al. Biochim Biophys Acta. 2009 Dec;1796(2):63-74. doi: 10.1016/j.bbcan.2009.02.002. Epub 2009 Mar 4. Biochim Biophys Acta. 2009. PMID: 19268509 Review.
Cited by
- FoxO proteins in the nervous system.
Maiese K. Maiese K. Anal Cell Pathol (Amst). 2015;2015:569392. doi: 10.1155/2015/569392. Epub 2015 Jun 10. Anal Cell Pathol (Amst). 2015. PMID: 26171319 Free PMC article. Review. - Erythropoietin and diabetes mellitus.
Maiese K. Maiese K. World J Diabetes. 2015 Oct 25;6(14):1259-73. doi: 10.4239/wjd.v6.i14.1259. World J Diabetes. 2015. PMID: 26516410 Free PMC article. Review. - Forkhead Transcription Factors: Formulating a FOXO Target for Cognitive Loss.
Maiese K. Maiese K. Curr Neurovasc Res. 2017;14(4):415-420. doi: 10.2174/1567202614666171116102911. Curr Neurovasc Res. 2017. PMID: 29149835 Free PMC article. Review. - FOXO3a governs early and late apoptotic endothelial programs during elevated glucose through mitochondrial and caspase signaling.
Hou J, Chong ZZ, Shang YC, Maiese K. Hou J, et al. Mol Cell Endocrinol. 2010 Jun 10;321(2):194-206. doi: 10.1016/j.mce.2010.02.037. Epub 2010 Mar 6. Mol Cell Endocrinol. 2010. PMID: 20211690 Free PMC article. - FOXO3a inhibits TNF-α- and IL-1β-induced astrocyte proliferation:Implication for reactive astrogliosis.
Cui M, Huang Y, Tian C, Zhao Y, Zheng J. Cui M, et al. Glia. 2011 Apr;59(4):641-54. doi: 10.1002/glia.21134. Epub 2011 Feb 3. Glia. 2011. PMID: 21294163 Free PMC article.
References
- Weigel D, Jurgens G, Kuttner F, et al. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the drosophila embryo. Cell. 1989;57(4):645–658. - PubMed
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14(2):142–146. - PubMed
- Parry P, Wei Y, Evans G. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 1994;11(2):79–84. - PubMed
- Hillion J, Le Coniat M, Jonveaux P, et al. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood. 1997;90(9):3714–3719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS053946-05/NS/NINDS NIH HHS/United States
- R01 NS053946-04/NS/NINDS NIH HHS/United States
- P30 ES06639/ES/NIEHS NIH HHS/United States
- R01 NS053946-01A2/NS/NINDS NIH HHS/United States
- R01 NS053946-03S1/NS/NINDS NIH HHS/United States
- R01 NS053946-03/NS/NINDS NIH HHS/United States
- P30 ES006639/ES/NIEHS NIH HHS/United States
- R01 NS053946-06/NS/NINDS NIH HHS/United States
- R01 NS053946/NS/NINDS NIH HHS/United States
- R01 NS053946-05W1/NS/NINDS NIH HHS/United States
- R01 NS053946-02/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous