PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach - PubMed (original) (raw)
. 2010 Jul;38(Web Server issue):W609-14.
doi: 10.1093/nar/gkq300. Epub 2010 Apr 29.
Affiliations
- PMID: 20430828
- PMCID: PMC2896160
- DOI: 10.1093/nar/gkq300
PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach
Xiaofeng Liu et al. Nucleic Acids Res. 2010 Jul.
Abstract
In silico drug target identification, which includes many distinct algorithms for finding disease genes and proteins, is the first step in the drug discovery pipeline. When the 3D structures of the targets are available, the problem of target identification is usually converted to finding the best interaction mode between the potential target candidates and small molecule probes. Pharmacophore, which is the spatial arrangement of features essential for a molecule to interact with a specific target receptor, is an alternative method for achieving this goal apart from molecular docking method. PharmMapper server is a freely accessed web server designed to identify potential target candidates for the given small molecules (drugs, natural products or other newly discovered compounds with unidentified binding targets) using pharmacophore mapping approach. PharmMapper hosts a large, in-house repertoire of pharmacophore database (namely PharmTargetDB) annotated from all the targets information in TargetBank, BindingDB, DrugBank and potential drug target database, including over 7000 receptor-based pharmacophore models (covering over 1500 drug targets information). PharmMapper automatically finds the best mapping poses of the query molecule against all the pharmacophore models in PharmTargetDB and lists the top N best-fitted hits with appropriate target annotations, as well as respective molecule's aligned poses are presented. Benefited from the highly efficient and robust triangle hashing mapping method, PharmMapper bears high throughput ability and only costs 1 h averagely to screen the whole PharmTargetDB. The protocol was successful in finding the proper targets among the top 300 pharmacophore candidates in the retrospective benchmarking test of tamoxifen. PharmMapper is available at http://59.78.96.61/pharmmapper.
Figures
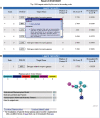
Figure 1.
An example of the output of PharmMapper. (A) The ranked list of hit target pharmacophore models, which are sorted by fit score in descending order. (B) The pull-down window that illustrates the details of each pharmacophore model candidate and the molecule pharmacophore alignment.
Similar articles
- PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database.
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J, Li H. Wang X, et al. Nucleic Acids Res. 2017 Jul 3;45(W1):W356-W360. doi: 10.1093/nar/gkx374. Nucleic Acids Res. 2017. PMID: 28472422 Free PMC article. - TarFisDock: a web server for identifying drug targets with docking approach.
Li H, Gao Z, Kang L, Zhang H, Yang K, Yu K, Luo X, Zhu W, Chen K, Shen J, Wang X, Jiang H. Li H, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W219-24. doi: 10.1093/nar/gkl114. Nucleic Acids Res. 2006. PMID: 16844997 Free PMC article. - PharmaGist: a webserver for ligand-based pharmacophore detection.
Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. Schneidman-Duhovny D, et al. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W223-8. doi: 10.1093/nar/gkn187. Epub 2008 Apr 19. Nucleic Acids Res. 2008. PMID: 18424800 Free PMC article. - Reverse Screening Methods to Search for the Protein Targets of Chemopreventive Compounds.
Huang H, Zhang G, Zhou Y, Lin C, Chen S, Lin Y, Mai S, Huang Z. Huang H, et al. Front Chem. 2018 May 9;6:138. doi: 10.3389/fchem.2018.00138. eCollection 2018. Front Chem. 2018. PMID: 29868550 Free PMC article. Review. - Virtual screening for the discovery of bioactive natural products.
Rollinger JM, Stuppner H, Langer T. Rollinger JM, et al. Prog Drug Res. 2008;65:211, 213-49. doi: 10.1007/978-3-7643-8117-2_6. Prog Drug Res. 2008. PMID: 18084917 Free PMC article. Review.
Cited by
- The Underlying Molecular Mechanisms Involved in Traditional Chinese Medicine Smilax china L. for the Treatment of Pelvic Inflammatory Disease.
Zhang Y, Zhao Z, Chen H, Fu Y, Wang W, Li Q, Li X, Wang X, Fan G, Zhang Y. Zhang Y, et al. Evid Based Complement Alternat Med. 2021 Apr 8;2021:5552532. doi: 10.1155/2021/5552532. eCollection 2021. Evid Based Complement Alternat Med. 2021. PMID: 33927774 Free PMC article. - Antitumor Activity and Mechanism of Robustic Acid from Dalbergia benthami Prain via Computational Target Fishing.
Huang J, Liang Y, Tian W, Ma J, Huang L, Li B, Chen R, Li D. Huang J, et al. Molecules. 2020 Aug 27;25(17):3919. doi: 10.3390/molecules25173919. Molecules. 2020. PMID: 32867345 Free PMC article. - Life beyond kinases: structure-based discovery of sorafenib as nanomolar antagonist of 5-HT receptors.
Lin X, Huang XP, Chen G, Whaley R, Peng S, Wang Y, Zhang G, Wang SX, Wang S, Roth BL, Huang N. Lin X, et al. J Med Chem. 2012 Jun 28;55(12):5749-59. doi: 10.1021/jm300338m. Epub 2012 Jun 19. J Med Chem. 2012. PMID: 22694093 Free PMC article. - Bioactive components and the molecular mechanism of Shengxian Decoction against lung adenocarcinoma based on network pharmacology and molecular docking.
Yuan R, Li K, Li Q, Wang C, Zhang H, Ge L, Ren Y, You F. Yuan R, et al. Am J Transl Res. 2023 Dec 15;15(12):6988-7012. eCollection 2023. Am J Transl Res. 2023. PMID: 38186989 Free PMC article. - Solasonine Induces Apoptosis and Inhibits Proliferation of Bladder Cancer Cells by Suppressing NRP1 Expression.
Dong Y, Hao L, Shi ZD, Fang K, Yu H, Zang GH, Fan T, Han CH. Dong Y, et al. J Oncol. 2022 Mar 2;2022:7261486. doi: 10.1155/2022/7261486. eCollection 2022. J Oncol. 2022. PMID: 35281516 Free PMC article.
References
- Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. - PubMed
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. - PubMed
- Bajorath J. Computational analysis of ligand relationships within target families. Curr. Opin. Chem. Biol. 2008;12:352–358. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical