The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation - PubMed (original) (raw)
doi: 10.1038/ni.1867. Epub 2010 May 2.
Sarita Sehra, Ritobrata Goswami, Weiguo Yao, Qing Yu, Gretta L Stritesky, Rukhsana Jabeen, Carl McKinley, Ayele-Nati Ahyi, Ling Han, Evelyn T Nguyen, Michael J Robertson, Narayanan B Perumal, Robert S Tepper, Stephen L Nutt, Mark H Kaplan
Affiliations
- PMID: 20431622
- PMCID: PMC3136246
- DOI: 10.1038/ni.1867
The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation
Hua-Chen Chang et al. Nat Immunol. 2010 Jun.
Abstract
CD4(+) helper T cells acquire effector phenotypes that promote specialized inflammatory responses. We show that the ETS-family transcription factor PU.1 was required for the development of an interleukin 9 (IL-9)-secreting subset of helper T cells. Decreasing PU.1 expression either by conditional deletion in mouse T cells or the use of small interfering RNA in human T cells impaired IL-9 production, whereas ectopic PU.1 expression promoted IL-9 production. Mice with PU.1-deficient T cells developed normal T helper type 2 (T(H)2) responses in vivo but showed attenuated allergic pulmonary inflammation that corresponded to lower expression of Il9 and chemokines in peripheral T cells and in lungs than that of wild-type mice. Together our data suggest a critical role for PU.1 in generating the IL-9-producing (T(H)9) phenotype and in the development of allergic inflammation.
Figures
Figure 1
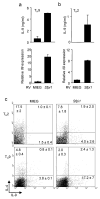
PU.1 is required for optimal IL-9 production in murine T cells. Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions. (a) After five days of culture, cells were stimulated with anti-CD3 and supernatants after 1, 2 or 3 days of stimulation were tested for IL-9 using ELISA. (b) RNA was isolated from wild-type and _Sfpi1_lck−/− TH2 and TH9 cultures after stimulation with anti-CD3. Il9 mRNA was assessed using qPCR. (c) Wild-type and _Sfpi1_lck−/– TH9 cultures were stimulated with PMA-ionomycin for 5 h before intracellular staining for IL-9. Results in (a-c) are the average of 3 mice and representative of more than four experiments. (d) Naïve CD4+ T cells cultured under TH1, TH2, TH9 or TH17 conditions were analyzed for Sfpi1 expression using qPCR. Results are an average of 3-5 experiments. (e) Naïve CD4+ T cells were cultured under TH9 or iTreg conditions for five days and stimulated with anti-CD3 for 6 h before intracellular staining for IL-9 and Foxp3. (f,g) Naïve CD4+ T cells were cultured under TH9 or TH17 conditions for five days and stimulated with (f) PMA-ionomycin for 6 h before intracellular staining for IL-9 and IL-17 or (g) anti-CD3 for 24 h before supernatants were collected for analysis using ELISA. Results in (e-g) are representative of at least three experiments. (h) TH2 cultures were separated into IL-4lo and IL-4hi populations using magnetic selection. RNA was isolated from separated cells to examine expression of Il4, Il9 and Sfpi1. Results are an average of three experiments. *, P < 0.05 using two-tailed student t test.
Figure 2
PU.1 promotes IL-9 production. TH9 (a,c) or TH2 (b,c) cultures were transduced with control (MIEG) or PU.1-expressing retroviruses. (a,b) EGFP-positive cells were sorted and stimulated to determine secretion of IL-9 (top) and Il9 expression (bottom). (c) IL-4 and IL-9 production was determined by intracellular staining in the EGFP+ populations of control and PU.1-transduced cells. Percentages in each quadrant are the average of results from 2-3 mice. Data are representative of 2-3 experiments.
Figure 3
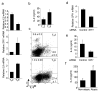
IL-9 and IL-10 are not coordinately regulated in TH9 cells. (a) Naïve CD4+ T cells were cultured under TH2 or TH9 conditions for five days before cells were left unstimulated or stimulated with plate-bound anti-CD3 for 6 or 24 hours and RNA was isolated for qPCR analysis of Il9 and Il10. (b) TH2 and TH9 cultures derived as in a were stimulated for 5 hours before intracellular cytokine analysis of IL-10 and IL-9. (c) Naïve CD4+ T cells were cultured under TH9 conditions in the presence or absence of anti-IL-10. After five days in culture cells were restimulated with anti-CD3 and supernatants were analyzed for cytokines using ELISA. (d) Naïve CD4+ T cells were cultured under TH9 conditions. After five days in culture, cells were restimulated with anti-CD3 in the presence or absence of TGF-β1 and supernatants were analyzed for cytokines using ELISA. (e) Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions for five days, and stimulated with anti-CD3 for 24 hours before supernatants were tested for IL-10 using ELISA. Results are representative of at least three experiments.
Figure 4
Histone modifications at the Il9 locus. (a) VISTA plot of conserved non-coding sequences adjacent to the Il9 locus. (b,c) Naïve CD4+ T cells were isolated and analyzed directly or differentiated under TH1, TH2, TH9, TH17 or iTreg culture conditions for five days. Cells were analyzed for histone modifications using chromatin immunoprecipitation for the indicated histone modifications and primers for Il9 CNS. (d,e) Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions and used for ChIP analysis as described in b performed for AcH3 at CNS1 and CNS2 (d), or for AcH4 and me3H3K27 at CNS1 (e). (f) Top, consensus PU.1 binding sites in the Il9 CNS1. Bottom, DAPA analysis of PU.1 binding from TH2 or TH9 extracts to site 2. For competition, extracts were incubated with a 5-fold excess of unlabelled PU.1 consensus double-stranded oligonucleotide. Expression of PU.1 in each cell extract is shown as input. Results are representative of three experiments. (g) Naïve CD4+ T cells were differentiated under TH1, TH2, or TH9 culture conditions for five days. Cells were analyzed for PU.1 binding by ChIP with primers for _Il9_CNS1. Results of ChIP experiments are shown as average ± SD of percent input with control IgG subtracted (a-e) or with anti-PU.1 and IgG control ChIP values shown separately (g) and are representative of 2-4 experiments.
Figure 5
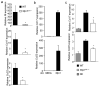
PU.1 promotes IL-9 production in human T cells. (a) Naïve human CD4+ T cells were cultured under TH2 or TH9 conditions and after five days in culture restimulated to analyze gene expression by qPCR. (b,c) TH2 and TH9 cultures were derived as in a and b supernatants from cells stimulated for 24 hours were tested for IL-9 production using ELISA or (c) cells were stimulated with anti-CD3 for 6 hours for analysis of IL-9 and IL-13 by intracellular cytokine staining. (d,e) Th9 cultures derived as in a were transfected with control or _SPI1_-specific siRNA. Twenty-four hours after transfection cells were analyzed for SPI1 expression by qPCR (d) and IL-9 production using ELISA (e). Results in (a-e) are representative data from 3-5 experiments with different donors. (f) PBMCs from infants (age 18-30 months) classified as atopic (n= 49) or non-atopic (n= 33) based on positive allergen-specific serum IgE, were stimulated with anti-CD3 for 48 hours and supernatants were tested for production of IL-9 using multiplex analysis. *, p<0.04 using an unpaired student t test.
Figure 6
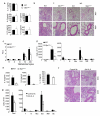
PU.1 expression in T cells is required for the development of allergic inflammation. Wild-type and _Sfpi1_lck−/− mice were sensitized and challenged intranasally with ovalbumin. Forty-eight hours after the last intranasal challenge mice were sacrificed for analysis. (a) Splenocytes were stimulated with anti-CD3 (left) or Ova (right) for 72 h and supernatants were tested for the amounts of TH2 cytokines by ELISA. (b) Lungs of challenged (C) or non-challenged (NC) mice were embedded in paraffin and analyzed by H/E staining. Magnification is indicated in the panel. (c) Wild-type and _Sfpi1_lck−/− mice were analyzed for airway hyperreactivity using whole body plethysmography 24 h after the last intranasal challenge. Airway function was tested at baseline (B), with saline inhalation (S) and following inhalation of the indicated doses of methacholine. (d) Numbers of cells recovered by bronchoalveolar lavage in challenged or non-challenged mice (left). Numbers of specific cell types in BAL from challenged mice, as determined by flow cytometry, are indicated (right). (e) Amounts of IL-9 and TH2 cytokines present in BAL fluid were determined using ELISA. (f, g) Wild type mice were sensitized and challenged intranasally with ovalbumin daily for five days. Mice were injected intravenously with control Ig or anti-IL-9, 30 minutes before the first, third and fifth intranasal challenge. Forty-eight hours after the last intranasal challenge, lungs were processed for histological analysis (f) as in b, and BAL cells were harvested and analyzed (g) as in d. *, p<0.05 using two-tailed student t test. All results are the average ± SEM of 5-7 mice and are representative of three experiments.
Figure 7
PU.1 is required for IL-9 and chemokine expression in allergen sensitized mice. (a) RNA isolated from Ova-stimulated splenocytes as described in (Fig. 6a) was analyzed for expression of Il9 and chemokines associated with allergic inflammation. (b) TH9 cultures were transduced with control (MIEG) or PU.1 expressing retroviruses. EGFP-positive cells were sorted and stimulated to determine expression of Ccl17 and Ccl22. (c) Total lung RNA was isolated from sensitized and challenged WT and _Sfpi1_lck−/− mice, or WT non-challenged (NC) mice, and analyzed for Ccl17 and Ccl22 mRNA by qPCR. *, p<0.05 using two-tailed student t test.
Comment in
- T cell responses: PU.1 in time saves nine.
Bordon Y. Bordon Y. Nat Rev Immunol. 2010 Jun;10(6):380. doi: 10.1038/nri2793. Nat Rev Immunol. 2010. PMID: 20514670 No abstract available.
Similar articles
- TH9 cells are required for tissue mast cell accumulation during allergic inflammation.
Sehra S, Yao W, Nguyen ET, Glosson-Byers NL, Akhtar N, Zhou B, Kaplan MH. Sehra S, et al. J Allergy Clin Immunol. 2015 Aug;136(2):433-40.e1. doi: 10.1016/j.jaci.2015.01.021. Epub 2015 Mar 5. J Allergy Clin Immunol. 2015. PMID: 25746972 Free PMC article. - The ETS Family Transcription Factors Etv5 and PU.1 Function in Parallel To Promote Th9 Cell Development.
Koh B, Hufford MM, Pham D, Olson MR, Wu T, Jabeen R, Sun X, Kaplan MH. Koh B, et al. J Immunol. 2016 Sep 15;197(6):2465-72. doi: 10.4049/jimmunol.1502383. Epub 2016 Aug 5. J Immunol. 2016. PMID: 27496971 Free PMC article. - p-STAT6, PU.1, and NF-κB are involved in allergen-induced late-phase airway inflammation in asthma patients.
Hoppenot D, Malakauskas K, Lavinskiene S, Sakalauskas R. Hoppenot D, et al. BMC Pulm Med. 2015 Oct 14;15:122. doi: 10.1186/s12890-015-0119-7. BMC Pulm Med. 2015. PMID: 26466682 Free PMC article. - T Helper 9 Cells: A New Player in Immune-Related Diseases.
Chen J, Guan L, Tang L, Liu S, Zhou Y, Chen C, He Z, Xu L. Chen J, et al. DNA Cell Biol. 2019 Oct;38(10):1040-1047. doi: 10.1089/dna.2019.4729. Epub 2019 Aug 16. DNA Cell Biol. 2019. PMID: 31414895 Free PMC article. Review. - Surprising new roles for PU.1 in the adaptive immune response.
Carotta S, Wu L, Nutt SL. Carotta S, et al. Immunol Rev. 2010 Nov;238(1):63-75. doi: 10.1111/j.1600-065X.2010.00955.x. Immunol Rev. 2010. PMID: 20969585 Review.
Cited by
- The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation.
Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, Kerketta R, Lee YH, Chang SH, Corry DB, Wang D, Watowich SS, Dong C. Yang XO, et al. Nat Immunol. 2013 Jul;14(7):732-40. doi: 10.1038/ni.2633. Epub 2013 Jun 2. Nat Immunol. 2013. PMID: 23727894 Free PMC article. - The Role of T Cells and Macrophages in Asthma Pathogenesis: A New Perspective on Mutual Crosstalk.
Zhu X, Cui J, Yi L, Qin J, Tulake W, Teng F, Tang W, Wei Y, Dong J. Zhu X, et al. Mediators Inflamm. 2020 Aug 19;2020:7835284. doi: 10.1155/2020/7835284. eCollection 2020. Mediators Inflamm. 2020. PMID: 32922208 Free PMC article. - Critical Roles of Balanced T Helper 9 Cells and Regulatory T Cells in Allergic Airway Inflammation and Tumor Immunity.
Huang M, Dong J. Huang M, et al. J Immunol Res. 2021 Mar 1;2021:8816055. doi: 10.1155/2021/8816055. eCollection 2021. J Immunol Res. 2021. PMID: 33748292 Free PMC article. Review. - Distinct chemokine receptor axes regulate Th9 cell trafficking to allergic and autoimmune inflammatory sites.
Kara EE, Comerford I, Bastow CR, Fenix KA, Litchfield W, Handel TM, McColl SR. Kara EE, et al. J Immunol. 2013 Aug 1;191(3):1110-7. doi: 10.4049/jimmunol.1203089. Epub 2013 Jun 24. J Immunol. 2013. PMID: 23797668 Free PMC article. - Recruitment and phenotypic characteristics of interleukin 9-producing CD4+ T cells in malignant pleural effusion.
Bu XN, Zhou Q, Zhang JC, Ye ZJ, Tong ZH, Shi HZ. Bu XN, et al. Lung. 2013 Aug;191(4):385-9. doi: 10.1007/s00408-013-9474-4. Epub 2013 May 23. Lung. 2013. PMID: 23700286
References
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. - PubMed
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. - PubMed
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. - PubMed
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI57459/AI/NIAID NIH HHS/United States
- U19 AI070448/AI/NIAID NIH HHS/United States
- T32 AI060519/AI/NIAID NIH HHS/United States
- R01 HL080071/HL/NHLBI NIH HHS/United States
- R01 CA118118/CA/NCI NIH HHS/United States
- R01 AI057459/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials