MicroRNAs control hepatocyte proliferation during liver regeneration - PubMed (original) (raw)
MicroRNAs control hepatocyte proliferation during liver regeneration
Guisheng Song et al. Hepatology. 2010 May.
Abstract
MicroRNAs (miRNAs) constitute a new class of regulators of gene expression. Among other actions, miRNAs have been shown to control cell proliferation in development and cancer. However, whether miRNAs regulate hepatocyte proliferation during liver regeneration is unknown. We addressed this question by performing 2/3 partial hepatectomy (2/3 PH) on mice with hepatocyte-specific inactivation of DiGeorge syndrome critical region gene 8 (DGCR8), an essential component of the miRNA processing pathway. Hepatocytes of these mice were miRNA-deficient and exhibited a delay in cell cycle progression involving the G(1) to S phase transition. Examination of livers of wildtype mice after 2/3 PH revealed differential expression of a subset of miRNAs, notably an induction of miR-21 and repression of miR-378. We further discovered that miR-21 directly inhibits Btg2, a cell cycle inhibitor that prevents activation of forkhead box M1 (FoxM1), which is essential for DNA synthesis in hepatocytes after 2/3 PH. In addition, we found that miR-378 directly inhibits ornithine decarboxylase (Odc1), which is known to promote DNA synthesis in hepatocytes after 2/3 PH.
Conclusion: Our results show that miRNAs are critical regulators of hepatocyte proliferation during liver regeneration. Because these miRNAs and target gene interactions are conserved, our findings may also be relevant to human liver regeneration.
Conflict of interest statement
Potential conflict of interest: Nothing to report.
Figures
Fig. 1
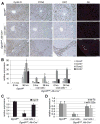
Global miRNA deficiency impairs G1 to S phase progression of hepatocytes after 2/3 PH and is accompanied by proliferation of wild-type oval cells. (A) Representative immunostainings show that hepatocytes lacking DGCR8 expression (hepatocytes of Dgcr8del/fl, Alb-Cre+/− mice) enter the G1 phase of the cell cycle (Cyclin D, brown) but, in contrast to hepatocytes of littermates with intact DGCR8 expression (hepatocytes of Dgcr8fl/fl mice), fail to progress into S phase (PCNA and Ki67, both brown) by 36 hours after 2/3 PH. However, by 72 hours after 2/3 PH hepatocytes of Dgcr8del/fl, Alb-Cre+/− mice progress into S phase and mitosis (Supporting Information Fig. 1A). A subset of Dgcr8del/fl, Alb-Cre+/− mice show proliferating oval cells (A6, red). These cells were detectable before 2/3 PH and retained normal DGCR8 expression (Supporting Information Fig. 2B). Oval cell activation is likely caused by the moderate hepatocyte injury detectable in Dgcr8del/fl, Alb-Cre+/− mice (Supporting Information Fig. 2C). (B) qRT-PCR 36 hours (hrs) after 2/3 PH shows that livers of Dgcr8del/fl, Alb-Cre+/− mice lacking oval cells fail to induce Ccna2 and Ccnb1, cyclins critical for DNA synthesis and mitosis. Livers from Dgcr8del/fl, Alb-Cre+/− mice containing oval cells show blunted induction of Ccna2 and Ccnb1 before and 36 hours after 2/3 PH. The difference in oval cell presence in mice of the same genotype is likely due to variable onset of Cre expression from the synthetic albumin promoter. Early Cre expression leads to DGCR8 inactivation in fetal liver progenitors which is inherited by hepatocytes, biliary cells, and oval cells. Cre activation after fetal liver progenitor lineage bifurcation restricts its expression to hepatocytes and leaves DGRC8 expression in oval cells intact. Thus, oval cells retain the ability to proliferate, whereas hepatocytes are negative for PCNA and Ki67 also in these mice at 36 hours after PH. Of note, mRNA levels of Ccnd1 are increased in Dgcr8del/fl, Alb-Cre+/− mice before 2/3 PH. (C) qRT-PCR shows loss of Dgcr8 expression in livers of Dgcr8del/fl, Alb-Cre+/− mice lacking oval cells. Because the synthetic albumin promoter restricts Cre expression to hepatocytes in adult mice, residual Dgcr8 expression in Dgcr8del/fl, Alb-Cre+/− mice lacking oval cells stems mostly from nonhepatocyte liver cell types. (D) Hepatocyte-specific Dgcr8 disruption leads to loss of the most abundant miRNAs in the liver, the hepatocyte-specific miR-122a, and, to a lesser extent, the ubiquitously expressed let-7b. miRNAs known to be processed independent of DGCR8 continue to be expressed in livers of Dgcr8del/fl, Alb-Cre+/− mice (Supporting Information Fig. 3A,B). Error bars represent ± SEM. *P < 0.005. Scale bars = 100 _μ_m.
Fig. 2
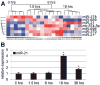
miRNA expression changes in livers of wildtype mice in response to 2/3 PH. (A) Heatmap of the miRNAs that are significantly differentially expressed in livers of wildtype mice during the first 18 hours after 2/3 PH. The miRNA clustering tree is shown on the left and the sample clustering tree appears at the top. The color scale in the bottom illustrates the relative expression level of an miRNA across all samples. Red color represents an expression level above mean, blue color represents expression lower than mean. The clustering is performed on log2 (Hy3/Hy5) ratios that passed the filtering criteria of P < 0.001 (Supporting Information Table 1). (B) qRT-PCR shows that expression of miR-21 peaks at 18 hours after 2/3 PH in wildtype mice. Error bars represent ± SEM. *P < 0.005.
Fig. 3
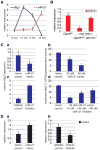
miR-21 directly antagonizes the proliferation-inhibiting gene Btg2 during liver regeneration. (A) qRT-PCR shows inverse correlation of Btg2 and miR-21 expression in the first 18 hours after 2/3 PH. miR-21 expression normalizes by 72 hours after 2/3 PH (Supporting Information Fig. 5A). (B) qRT-PCR shows loss of miR-21 in livers of Dgcr8del/fl, Alb-Cre+/− mice lacking oval cells. Livers of Dgcr8del/fl, Alb-Cre+/− mice containing oval cells have miR-21 levels similar to controls, reflecting the proliferative activity of oval cells. (C) miR-21 mimic transfection into Hepa1,6 cells causes a reduction in Btg2 mRNA levels (a) and dose-dependent inhibition of the activity of a luciferase reporter gene linked to the 3′UTR of Btg2 (b). Conversely, miR-21 inhibitor transfection into Hepa1,6 cells causes an increase in Btg2 mRNA levels (c). Furthermore, miR-21 inhibitor transfection antagonizes the inhibitory binding of miR-21 mimics to the 3′UTR of Btg2 (d). (D) Levels of Ccnb1 mRNA are increased after miR-21 mimic (a) and decreased after mir-21 inhibitor (b) transfection into Hepa1,6 cells. Error bars represent ± SEM. * P < 0.005. #P < 0.05.
Fig. 4
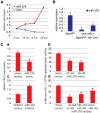
Declining expression of miR-378 during liver regeneration leads to de-repression of its proliferation-promoting target gene Odc1. (A) qRT-PCR shows inverse correlation of Odc1 and miR-378 expression in the first 18 hours after 2/3 PH. miR-378 expression normalizes by 72 hours after 2/3 PH (Supporting Information Fig. 5A). (B) qRT-PCR shows loss of miR-378 in livers of Dgcr8del/fl, Alb-Cre+/− mice lacking oval cells. Livers of Dgcr8del/fl, Alb-Cre+/− mice containing oval cells have significantly lower miR-378 levels than controls, suggesting that miR-378 expression increases with hepatic differentiation. (C) miR-378 mimic transfection into Hepa1,6 cells causes a reduction in Odc1 mRNA levels (a) and dose-dependent inhibition of the activity of a luciferase reporter gene linked to the 3′UTR of Odc1 (b). Conversely, miR-378 inhibitor transfection into Hepa1,6 cells causes an increase in Odc1 mRNA levels (c). Furthermore, miR-378 inhibitor transfection antagonizes the inhibitory binding of miR-378 mimics to the 3′UTR of Odc1 (d). Error bars represent ± SEM. *P < 0.005. ## P < 0.01. #P < 0.05.
Similar articles
- MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver.
Kim J, Hyun J, Wang S, Lee C, Jung Y. Kim J, et al. Cell Death Dis. 2018 Jun 18;9(7):721. doi: 10.1038/s41419-018-0762-z. Cell Death Dis. 2018. PMID: 29915286 Free PMC article. - Dicer-dependent production of microRNA221 in hepatocytes inhibits p27 and is required for liver regeneration in mice.
Oya Y, Masuzaki R, Tsugawa D, Ray KC, Dou Y, Karp SJ. Oya Y, et al. Am J Physiol Gastrointest Liver Physiol. 2017 May 1;312(5):G464-G473. doi: 10.1152/ajpgi.00383.2016. Epub 2017 Feb 23. Am J Physiol Gastrointest Liver Physiol. 2017. PMID: 28232457 Free PMC article. - Inhibition of miR-21 rescues liver regeneration after partial hepatectomy in ethanol-fed rats.
Juskeviciute E, Dippold RP, Antony AN, Swarup A, Vadigepalli R, Hoek JB. Juskeviciute E, et al. Am J Physiol Gastrointest Liver Physiol. 2016 Nov 1;311(5):G794-G806. doi: 10.1152/ajpgi.00292.2016. Epub 2016 Sep 15. Am J Physiol Gastrointest Liver Physiol. 2016. PMID: 27634014 Free PMC article. - MicroRNAs in Liver Regeneration.
Chen X, Zhao Y, Wang F, Bei Y, Xiao J, Yang C. Chen X, et al. Cell Physiol Biochem. 2015;37(2):615-28. doi: 10.1159/000430381. Cell Physiol Biochem. 2015. PMID: 26344368 Review. - The role of microRNAs in the different phases of liver regeneration.
Kern AE, Ortmayr G, Assinger A, Starlinger P. Kern AE, et al. Expert Rev Gastroenterol Hepatol. 2023 Jul-Dec;17(10):959-973. doi: 10.1080/17474124.2023.2267422. Epub 2023 Oct 24. Expert Rev Gastroenterol Hepatol. 2023. PMID: 37811642 Review.
Cited by
- A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration.
Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. Ng R, et al. J Clin Invest. 2012 Mar;122(3):1097-108. doi: 10.1172/JCI46039. Epub 2012 Feb 13. J Clin Invest. 2012. PMID: 22326957 Free PMC article. - MicroRNA-124 and microRNA-378 inhibit the proliferation and invasion of colorectal cancer by upregulating KiSS1.
Zheng Y, Liu Y, Lin Y, Lin S, Gao J, Chen Z, Chen S. Zheng Y, et al. Transl Cancer Res. 2020 Apr;9(4):2838-2846. doi: 10.21037/tcr.2020.02.30. Transl Cancer Res. 2020. PMID: 35117640 Free PMC article. - miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis.
Dongiovanni P, Meroni M, Longo M, Fargion S, Fracanzani AL. Dongiovanni P, et al. Int J Mol Sci. 2018 Dec 10;19(12):3966. doi: 10.3390/ijms19123966. Int J Mol Sci. 2018. PMID: 30544653 Free PMC article. Review. - MicroRNA-194 protects against chronic hepatitis B-related liver damage by promoting hepatocyte growth via ACVR2B.
Gao X, Zhao P, Hu J, Zhu H, Zhang J, Zhou Z, Zhao J, Tang F. Gao X, et al. J Cell Mol Med. 2018 Sep;22(9):4534-4544. doi: 10.1111/jcmm.13714. Epub 2018 Jul 25. J Cell Mol Med. 2018. PMID: 30044042 Free PMC article. - Dynamic microRNA profiles of hepatic differentiated human umbilical cord lining-derived mesenchymal stem cells.
Cui L, Zhou X, Li J, Wang L, Wang J, Li Q, Chu J, Zheng L, Wu Q, Han Z, Shi Y, Han Y, Fan D. Cui L, et al. PLoS One. 2012;7(9):e44737. doi: 10.1371/journal.pone.0044737. Epub 2012 Sep 12. PLoS One. 2012. PMID: 22984549 Free PMC article.
References
- Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous