Clinical assessment incorporating a personal genome - PubMed (original) (raw)
. 2010 May 1;375(9725):1525-35.
doi: 10.1016/S0140-6736(10)60452-7.
Atul J Butte, Matthew T Wheeler, Rong Chen, Teri E Klein, Frederick E Dewey, Joel T Dudley, Kelly E Ormond, Aleksandra Pavlovic, Alexander A Morgan, Dmitry Pushkarev, Norma F Neff, Louanne Hudgins, Li Gong, Laura M Hodges, Dorit S Berlin, Caroline F Thorn, Katrin Sangkuhl, Joan M Hebert, Mark Woon, Hersh Sagreiya, Ryan Whaley, Joshua W Knowles, Michael F Chou, Joseph V Thakuria, Abraham M Rosenbaum, Alexander Wait Zaranek, George M Church, Henry T Greely, Stephen R Quake, Russ B Altman
Affiliations
- PMID: 20435227
- PMCID: PMC2937184
- DOI: 10.1016/S0140-6736(10)60452-7
Clinical assessment incorporating a personal genome
Euan A Ashley et al. Lancet. 2010.
Abstract
Background: The cost of genomic information has fallen steeply, but the clinical translation of genetic risk estimates remains unclear. We aimed to undertake an integrated analysis of a complete human genome in a clinical context.
Methods: We assessed a patient with a family history of vascular disease and early sudden death. Clinical assessment included analysis of this patient's full genome sequence, risk prediction for coronary artery disease, screening for causes of sudden cardiac death, and genetic counselling. Genetic analysis included the development of novel methods for the integration of whole genome and clinical risk. Disease and risk analysis focused on prediction of genetic risk of variants associated with mendelian disease, recognised drug responses, and pathogenicity for novel variants. We queried disease-specific mutation databases and pharmacogenomics databases to identify genes and mutations with known associations with disease and drug response. We estimated post-test probabilities of disease by applying likelihood ratios derived from integration of multiple common variants to age-appropriate and sex-appropriate pre-test probabilities. We also accounted for gene-environment interactions and conditionally dependent risks.
Findings: Analysis of 2.6 million single nucleotide polymorphisms and 752 copy number variations showed increased genetic risk for myocardial infarction, type 2 diabetes, and some cancers. We discovered rare variants in three genes that are clinically associated with sudden cardiac death-TMEM43, DSP, and MYBPC3. A variant in LPA was consistent with a family history of coronary artery disease. The patient had a heterozygous null mutation in CYP2C19 suggesting probable clopidogrel resistance, several variants associated with a positive response to lipid-lowering therapy, and variants in CYP4F2 and VKORC1 that suggest he might have a low initial dosing requirement for warfarin. Many variants of uncertain importance were reported.
Interpretation: Although challenges remain, our results suggest that whole-genome sequencing can yield useful and clinically relevant information for individual patients.
Funding: National Institute of General Medical Sciences; National Heart, Lung And Blood Institute; National Human Genome Research Institute; Howard Hughes Medical Institute; National Library of Medicine, Lucile Packard Foundation for Children's Health; Hewlett Packard Foundation; Breetwor Family Foundation.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
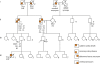
Figure 1
Patient pedigree. The arrow indicates the patient. Square shapes represent males, circles represent females. ARVD/C – arrhythmogenic right ventricular dysplasia/cardiomyopathy; AAA – abdominal aortic aneurysm;, HTN – hypertension; CAD – coronary artery disease; VT – paroxysmal ventricular tachycardia; HC – hypercholesterolemia; ARMD – age related macular degeneration; OA – osteoarthritis; SCD – sudden cardiac death (presumed)
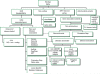
Figure 2
Approach to rare or novel variants See text for details. GVS - Genome Variation Server; SIFT - Sorting Intolerant From Tolerant; HGMD – Human Gene Mutation; Polyphen – POLYmorphism PHENotyping; HGVS – Human genome variation society; ARVC Database; mtSNP - mtSNP: a database of human mitochondrial genome polymorphisms; UniProt - UNIversal PROTein Resource; PolyDom – a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease; OMIM -Online Mendelian Inheritance in Man, OMIM.
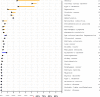
Figure 3A
Clinical risk incorporating genetic risk estimates for major diseases. Post-test probabilities were calculated by multiplying published pre-test probabilities or disease prevalence (in Caucasian males in the patient's age range, when available; see also Supplemental Table 4) with a series of independent likelihood ratios for each patient allele. Only the 32 diseases with (1) available pre-test probabilities, (2) more than one associated SNP, and (3) with published genotype frequencies are shown here. Disorders such as abdominal aortic aneurysm and progressive supranuclear palsy are not listed here, because they are diseases with only one available SNP. The back of the arrow heads indicate pre-test probabilities, and the point in the direction of the change in probability. Blue lines indicate a lowered post-test probability, and orange indicates an increased post-test probability. The number of independent SNPs used in the calculation of post-test probability for each disease is shown in the right. The advantage of plotting pre and post-test probabilities is illustrated by several cases. For example, While the patient has increased genetic risk for Graves' disease, the pre-test probability of this disease is very low so that post-test probability also remains low. However, while the patient exhibits much less genetic contribution to his risk for prostate cancer, his prior probability is high.
Figure 3B
The contribution of individual alleles to overall risk for four example diseases. Using our pre-test probability estimate as a starting point, we use SNPs with an association published from a genome wide association study, and then order them, as the number of studies showing association and sample sizes decrease. The darker colour indicates a greater number of published studies reporting association of that SNP with the disease, and the size of the box scales with the logarithm of the number of samples used to calculate the likelihood ratio. The SNP related gene if known, and the patient's genotype calls, are shown to the left of the diagram. The right shows, for each SNP, the likelihood ratio of the disease for the patient's genotype, the number of studies reporting an association, the number of samples used to calculate the likelihood ratio, and the post-test probability to that point down the graph. SNPs at the top of the graph are reported in more and larger studies, and we have greater confidence in their association with disease. The test probabilities are calculated by serially stepping down the list of SNPs and calculating an updated post-test probability using the contribution of that genotype, while including the contribution to our estimate of the SNPs above.
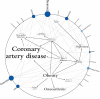
Figure 3C. Gene-environment interaction
A conditional dependency diagram for diseases represented in the patient's genetic risk profile. Only diseases for which a calculable post-test risk probability > 10% are shown. The size of the disease name text is proportional to the post-test risk probability. A solid black directed edge is drawn between disease names if one disease is known to be a predisposing aetiological factor for another disease. Environmental factors that are potentially modifiable are shown around the circumference, and dashed grey directed edge is drawn between an environmental factor and a disease if the factor has been frequently published in association with the aetiology of the disease. Environmental factors are portrayed in a size proportional to the number of diseases they are associated with in the circuit. The intensity of the colours of the factor circles represents the maximum post-test risk probability among those diseases directly associated with each factor.
Comment in
- The personal genome--the future of personalised medicine?
Samani NJ, Tomaszewski M, Schunkert H. Samani NJ, et al. Lancet. 2010 May 1;375(9725):1497-8. doi: 10.1016/S0140-6736(10)60598-3. Lancet. 2010. PMID: 20435212 No abstract available. - Clinical assessment incorporating a personal genome.
Pierce BL, Ahsan H. Pierce BL, et al. Lancet. 2010 Sep 11;376(9744):869; author reply 869-70. doi: 10.1016/S0140-6736(10)61404-3. Lancet. 2010. PMID: 20833292 No abstract available.
Similar articles
- A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose.
Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. Takeuchi F, et al. PLoS Genet. 2009 Mar;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. Epub 2009 Mar 20. PLoS Genet. 2009. PMID: 19300499 Free PMC article. - An evaluation of nine genetic variants related to metabolism and mechanism of action of warfarin as applied to stable dose prediction.
Carlquist JF, Horne BD, Mower C, Park J, Huntinghouse J, McKinney JT, Muhlestein JB, Anderson JL. Carlquist JF, et al. J Thromb Thrombolysis. 2010 Oct;30(3):358-64. doi: 10.1007/s11239-010-0467-3. J Thromb Thrombolysis. 2010. PMID: 20499136 - Dependency of phenprocoumon dosage on polymorphisms in the VKORC1, CYP2C9, and CYP4F2 genes.
Teichert M, Eijgelsheim M, Uitterlinden AG, Buhre PN, Hofman A, De Smet PA, Visser LE, Stricker BH. Teichert M, et al. Pharmacogenet Genomics. 2011 Jan;21(1):26-34. doi: 10.1097/FPC.0b013e32834154fb. Pharmacogenet Genomics. 2011. PMID: 21063236 - Clinical application of cardiovascular pharmacogenetics.
Voora D, Ginsburg GS. Voora D, et al. J Am Coll Cardiol. 2012 Jul 3;60(1):9-20. doi: 10.1016/j.jacc.2012.01.067. J Am Coll Cardiol. 2012. PMID: 22742397 Review. - Emerging genomic applications in coronary artery disease.
Damani SB, Topol EJ. Damani SB, et al. JACC Cardiovasc Interv. 2011 May;4(5):473-82. doi: 10.1016/j.jcin.2010.12.016. JACC Cardiovasc Interv. 2011. PMID: 21596318 Free PMC article. Review.
Cited by
- Chapter 7: Pharmacogenomics.
Karczewski KJ, Daneshjou R, Altman RB. Karczewski KJ, et al. PLoS Comput Biol. 2012;8(12):e1002817. doi: 10.1371/journal.pcbi.1002817. Epub 2012 Dec 27. PLoS Comput Biol. 2012. PMID: 23300409 Free PMC article. - Incidental copy-number variants identified by routine genome testing in a clinical population.
Boone PM, Soens ZT, Campbell IM, Stankiewicz P, Cheung SW, Patel A, Beaudet AL, Plon SE, Shaw CA, McGuire AL, Lupski JR. Boone PM, et al. Genet Med. 2013 Jan;15(1):45-54. doi: 10.1038/gim.2012.95. Epub 2012 Aug 9. Genet Med. 2013. PMID: 22878507 Free PMC article. - Parent and public interest in whole-genome sequencing.
Dodson DS, Goldenberg AJ, Davis MM, Singer DC, Tarini BA. Dodson DS, et al. Public Health Genomics. 2015;18(3):151-9. doi: 10.1159/000375115. Epub 2015 Mar 6. Public Health Genomics. 2015. PMID: 25765282 Free PMC article. - Next generation sequencing in clinical medicine: Challenges and lessons for pathology and biomedical informatics.
Gullapalli RR, Desai KV, Santana-Santos L, Kant JA, Becich MJ. Gullapalli RR, et al. J Pathol Inform. 2012;3:40. doi: 10.4103/2153-3539.103013. Epub 2012 Oct 31. J Pathol Inform. 2012. PMID: 23248761 Free PMC article. - Molecular genetic testing and the future of clinical genomics.
Katsanis SH, Katsanis N. Katsanis SH, et al. Nat Rev Genet. 2013 Jun;14(6):415-26. doi: 10.1038/nrg3493. Nat Rev Genet. 2013. PMID: 23681062 Free PMC article. Review.
References
- Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 GM061374/GM/NIGMS NIH HHS/United States
- T15 LM007033-23/LM/NLM NIH HHS/United States
- K08 HL083914-04/HL/NHLBI NIH HHS/United States
- F32HL097462/HL/NHLBI NIH HHS/United States
- GM61374/GM/NIGMS NIH HHS/United States
- U01 GM061374/GM/NIGMS NIH HHS/United States
- R01 GM079719-04/GM/NIGMS NIH HHS/United States
- R01 GM079719/GM/NIGMS NIH HHS/United States
- F32 HL097462-01/HL/NHLBI NIH HHS/United States
- GM079719/GM/NIGMS NIH HHS/United States
- U01 GM061374-07/GM/NIGMS NIH HHS/United States
- HG003389/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 LM009719-02/LM/NLM NIH HHS/United States
- K08 HL083914/HL/NHLBI NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- F32 HL097462/HL/NHLBI NIH HHS/United States
- P50 HG003389/HG/NHGRI NIH HHS/United States
- T32 HL094274/HL/NHLBI NIH HHS/United States
- P50 HG003389-05/HG/NHGRI NIH HHS/United States
- LM009719/LM/NLM NIH HHS/United States
- R01 LM009719/LM/NLM NIH HHS/United States
- LM007033/LM/NLM NIH HHS/United States
- DP2 OD006511/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous