Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation - PubMed (original) (raw)
Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation
Cole Trapnell et al. Nat Biotechnol. 2010 May.
Abstract
High-throughput mRNA sequencing (RNA-Seq) promises simultaneous transcript discovery and abundance estimation. However, this would require algorithms that are not restricted by prior gene annotations and that account for alternative transcription and splicing. Here we introduce such algorithms in an open-source software program called Cufflinks. To test Cufflinks, we sequenced and analyzed >430 million paired 75-bp RNA-Seq reads from a mouse myoblast cell line over a differentiation time series. We detected 13,692 known transcripts and 3,724 previously unannotated ones, 62% of which are supported by independent expression data or by homologous genes in other species. Over the time series, 330 genes showed complete switches in the dominant transcription start site (TSS) or splice isoform, and we observed more subtle shifts in 1,304 other genes. These results suggest that Cufflinks can illuminate the substantial regulatory flexibility and complexity in even this well-studied model of muscle development and that it can improve transcriptome-based genome annotation.
Figures
Figure 1
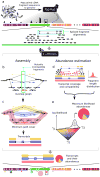
Overview of Cufflinks. The algorithm takes as input cDNA fragment sequences that have been (a) aligned to the genome by software capable of producing spliced alignments, such as TopHat. With paired-end RNA-Seq, Cufflinks treats each pair of fragment reads as a single alignment. The algorithm assembles overlapping ‘bundles’ of fragment alignments (b-c) separately, which reduces running time and memory use because each bundle typically contains the fragments from no more than a few genes. Cufflinks then estimates the abundances of the assembled transcripts (d-e). (b) The first step in fragment assembly is to identify pairs of ‘incompatible’ fragments that must have originated from distinct spliced mRNA isoforms. Fragments are connected in an ‘overlap graph’ when they are compatible and their alignments overlap in the genome. Each fragment has one node in the graph, and an edge, directed from left to right along the genome, is placed between each pair of compatible fragments. In this example, the yellow, blue, and red fragments must have originated from separate isoforms, but any other fragment could have come from the same transcript as one of these three. (c) Assembling isoforms from the overlap graph. Paths through the graph correspond to sets of mutually compatible fragments that could be merged into complete isoforms. The overlap graph here can be minimally ‘covered’ by three paths, each representing a different isoform. Dilworth's Theorem states that the number of mutually incompatible reads is the same as the minimum number of transcripts needed to “explain” all the fragments. Cufflinks implements a proof of Dilworth's Theorem that produces a minimal set of paths that cover all the fragments in the overlap graph by finding the largest set of reads with the property that no two could have originated from the same isoform. (d) Estimating transcript abundance. Fragments are matched (denoted here using color) to the transcripts from which they could have originated. The violet fragment could have originated from the blue or red isoform. Gray fragments could have come from any of the three shown. Cufflinks estimates transcript abundances using a statistical model in which the probability of observing each fragment is a linear function of the abundances of the transcripts from which it could have originated. Because only the ends of each fragment are sequenced, the length of each may be unknown. Assigning a fragment to different isoforms often implies a different length for it. Cufflinks can incorporate the distribution of fragment lengths to help assign fragments to isoforms. For example, the violet fragment would be much longer, and very improbable according to Cufflinks' model, if it were to come from the red isoform instead of the blue isoform. (e) The program then numerically maximizes a function that assigns a likelihood to all possible sets of relative abundances of the yellow, red and blue isoforms (γ1,γ2,γ3), producing the abundances that best explain the observed fragments, shown as a pie chart.
Figure 2
Distinction of transcriptional and post-transcriptional regulatory effects on overall transcript output. (a) When abundances of isoforms A, B, and C of Myc are grouped by TSS, changes in the relative abundances of the TSS groups indicate transcriptional regulation. Post-transcriptional effects are seen in changes in levels of isoforms of a single TSS group. (b) Isoforms of Myc have distinct expression dynamics. (c) Myc isoforms are downregulated as the time course proceeds. The width of the colored band is the measure of change in relative transcript abundance and the color is the log ratio of transcriptional and post-transcriptional contributions to change in relative abundances (plot construction detailed in Supplementary Method Section 5.3). Changes in relative abundances of Myc isoforms suggest that transcriptional effects immediately following differentiation at 0 hours give way to post-transcriptional effects later in the time course, as isoform A is eliminated.
Figure 2
Distinction of transcriptional and post-transcriptional regulatory effects on overall transcript output. (a) When abundances of isoforms A, B, and C of Myc are grouped by TSS, changes in the relative abundances of the TSS groups indicate transcriptional regulation. Post-transcriptional effects are seen in changes in levels of isoforms of a single TSS group. (b) Isoforms of Myc have distinct expression dynamics. (c) Myc isoforms are downregulated as the time course proceeds. The width of the colored band is the measure of change in relative transcript abundance and the color is the log ratio of transcriptional and post-transcriptional contributions to change in relative abundances (plot construction detailed in Supplementary Method Section 5.3). Changes in relative abundances of Myc isoforms suggest that transcriptional effects immediately following differentiation at 0 hours give way to post-transcriptional effects later in the time course, as isoform A is eliminated.
Figure 2
Distinction of transcriptional and post-transcriptional regulatory effects on overall transcript output. (a) When abundances of isoforms A, B, and C of Myc are grouped by TSS, changes in the relative abundances of the TSS groups indicate transcriptional regulation. Post-transcriptional effects are seen in changes in levels of isoforms of a single TSS group. (b) Isoforms of Myc have distinct expression dynamics. (c) Myc isoforms are downregulated as the time course proceeds. The width of the colored band is the measure of change in relative transcript abundance and the color is the log ratio of transcriptional and post-transcriptional contributions to change in relative abundances (plot construction detailed in Supplementary Method Section 5.3). Changes in relative abundances of Myc isoforms suggest that transcriptional effects immediately following differentiation at 0 hours give way to post-transcriptional effects later in the time course, as isoform A is eliminated.
Figure 3
Excluding isoforms discovered by Cufflinks from the transcript abundance estimation impacts the abundance estimates of known isoforms, in some cases by orders of magnitude. Four-and-a-half-LIM domains 3 (Fhl3) inhibits myogenesis by binding MyoD and attenuating its transcriptional activity. (a) The C2C12 transcriptome contains a novel isoform that is dominant during proliferation. The new TSS for Fhl3 is supported by proximal TAF1 and RNA polymerase II ChIP-Seq peaks. (b) The known isoform (solid line) is preferred at time points following differentiation.
Figure 3
Excluding isoforms discovered by Cufflinks from the transcript abundance estimation impacts the abundance estimates of known isoforms, in some cases by orders of magnitude. Four-and-a-half-LIM domains 3 (Fhl3) inhibits myogenesis by binding MyoD and attenuating its transcriptional activity. (a) The C2C12 transcriptome contains a novel isoform that is dominant during proliferation. The new TSS for Fhl3 is supported by proximal TAF1 and RNA polymerase II ChIP-Seq peaks. (b) The known isoform (solid line) is preferred at time points following differentiation.
Figure 4
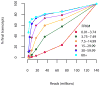
Robustness of assembly and abundance estimation as a function of expression level and depth of sequencing. Subsets of the full 60-hour read set were mapped and assembled with TopHat and Cufflinks and the resulting assemblies were compared for structural and abundance agreement with the full 60 hour assembly. Colored lines show the results obtained at different depths of sequencing in the full assembly; e.g., the light blue line tracks the performance for transcripts with FPKM greater than 60. (a) The fraction of transcript fragments fully recovered increases with additional sequencing data, though nearly 75% of moderately expressed (≥15 FPKM) are recovered with less than 40 million 75bp paired-end reads (20 million fragments), a fraction of the data generated by a single run of the sequencer used in this experiment. (b) Abundance estimates are similarly robust. At 40 million reads, transcripts determined to be moderately expressed using all 60 hour reads were estimated at within 15% of their final FPKM values.
Figure 4
Robustness of assembly and abundance estimation as a function of expression level and depth of sequencing. Subsets of the full 60-hour read set were mapped and assembled with TopHat and Cufflinks and the resulting assemblies were compared for structural and abundance agreement with the full 60 hour assembly. Colored lines show the results obtained at different depths of sequencing in the full assembly; e.g., the light blue line tracks the performance for transcripts with FPKM greater than 60. (a) The fraction of transcript fragments fully recovered increases with additional sequencing data, though nearly 75% of moderately expressed (≥15 FPKM) are recovered with less than 40 million 75bp paired-end reads (20 million fragments), a fraction of the data generated by a single run of the sequencer used in this experiment. (b) Abundance estimates are similarly robust. At 40 million reads, transcripts determined to be moderately expressed using all 60 hour reads were estimated at within 15% of their final FPKM values.
Comment in
- Advancing RNA-Seq analysis.
Haas BJ, Zody MC. Haas BJ, et al. Nat Biotechnol. 2010 May;28(5):421-3. doi: 10.1038/nbt0510-421. Nat Biotechnol. 2010. PMID: 20458303 No abstract available.
References
- Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5:613–619. -PubMed
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. -PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM083873/GM/NIGMS NIH HHS/United States
- R01-LM006845/LM/NLM NIH HHS/United States
- R01 HG006102/HG/NHGRI NIH HHS/United States
- R01 LM006845/LM/NLM NIH HHS/United States
- U54-HG004576/HG/NHGRI NIH HHS/United States
- U54 HG004576/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases