Computational challenges in the analysis of ancient DNA - PubMed (original) (raw)
Computational challenges in the analysis of ancient DNA
Kay Prüfer et al. Genome Biol. 2010.
Abstract
High-throughput sequencing technologies have opened up a new avenue for studying extinct organisms. Here we identify and quantify biases introduced by particular characteristics of ancient DNA samples. These analyses demonstrate the importance of closely related genomic sequence for correctly identifying and classifying bona fide endogenous DNA fragments. We show that more accurate genome divergence estimates from ancient DNA sequence can be attained using at least two outgroup genomes and appropriate filtering.
Figures
Figure 1
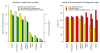
Number of aligned ancient DNA fragments and average sequence length. Properties of Mega BLAST alignments of ancient DNA sequences from a Neandertal fossil to genome sequences of increasing divergence. Left panel: number of reads with a best hit to the genome sequence and not to the GenBank nonredundant and environmental databases (yellow). Subset of reads with one unique best hit to the reference genome (light green). Subset of reads with one unique best hit to the reference genome that can be fully aligned with a positive alignment score (dark green). Right panel: Average length of best local alignments (yellow), average length of fragments with a unique best local alignment (red), average length of fragments with a positive score when fully aligned to reference genome (brown).
Figure 2
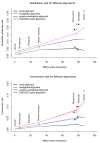
Differences per site in alignments of ancient DNA fragments. All nucleotide differences (top) and transversion differences (bottom) in different alignments to reference genomes of increasing divergence. Each read is required to have one uniquely best Mega BLAST alignment to the reference genome (estimate shown as the black line). The semiglobal alignment forces the full sequence to align to the genomic region identified by the local alignment (estimate shown as red line). These full alignments are further filtered for having a positive alignment score (blue line). The green crosses show the differences between human and the reference species in the ENCODE multiple sequence alignments. The divergence times on the x-axis are from [52] and [35], except for human for which we choose an arbitrary divergence time of 1 million years to Neandertal.
Figure 3
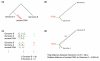
Schematic description of divergence triangulation. (a) A phylogenetic tree depicting the necessary topology for the application of the divergence triangulation method. (b) The ancient DNA sequences are used like an outgroup to the two genomic sequences in an unrooted tree. (c) Alignments between genomic sequences and ancient DNA fragments are used to assign changes to the lineages (numbers on the right-hand side). In this process, coinciding changes often caused by ancient DNA damage (shown in red in the alignments) can lead to misassignments of differences (in red in the summary of tables) (d) The assigned differences can be used to calculate a divergence relative to the divergence between the two genome sequences.
Figure 4
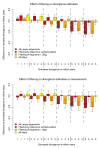
Divergence estimates by triangulation on simulated datasets. (a) 3DP divergence estimates in comparison to the expected values. Four bars are drawn for different filters: raw estimate without filtering on all unique alignments (brown); filtered alignments with verified human and chimpanzee genomic location using a whole genome alignment and a distance of at least 6 points between best and second best local alignments' bitscores (red); alignments of fragments with a size >35 bp (orange); and all filters applied (yellow). (b) Estimates are derived solely from transversion differences, otherwise identical to (a).
Similar articles
- Alignment-free estimation of nucleotide diversity.
Haubold B, Reed FA, Pfaffelhuber P. Haubold B, et al. Bioinformatics. 2011 Feb 15;27(4):449-55. doi: 10.1093/bioinformatics/btq689. Epub 2010 Dec 14. Bioinformatics. 2011. PMID: 21156730 - An Eulerian path approach to global multiple alignment for DNA sequences.
Zhang Y, Waterman MS. Zhang Y, et al. J Comput Biol. 2003;10(6):803-19. doi: 10.1089/106652703322756096. J Comput Biol. 2003. PMID: 14980012 - Comparative testing of DNA segmentation algorithms using benchmark simulations.
Elhaik E, Graur D, Josic K. Elhaik E, et al. Mol Biol Evol. 2010 May;27(5):1015-24. doi: 10.1093/molbev/msp307. Epub 2009 Dec 16. Mol Biol Evol. 2010. PMID: 20018981 - Mitogenomic analyses from ancient DNA.
Paijmans JL, Gilbert MT, Hofreiter M. Paijmans JL, et al. Mol Phylogenet Evol. 2013 Nov;69(2):404-16. doi: 10.1016/j.ympev.2012.06.002. Epub 2012 Jun 15. Mol Phylogenet Evol. 2013. PMID: 22705825 Review. - Revisiting Evaluation of Multiple Sequence Alignment Methods.
Warnow T. Warnow T. Methods Mol Biol. 2021;2231:299-317. doi: 10.1007/978-1-0716-1036-7_17. Methods Mol Biol. 2021. PMID: 33289899 Review.
Cited by
- SAFARI: Pangenome Alignment of Ancient DNA Using Purine/Pyrimidine Encodings.
Rubin J, van Waaij J, Kraft L, Sirén J, Sackett PW, Renaud G. Rubin J, et al. bioRxiv [Preprint]. 2024 Oct 8:2024.08.12.607489. doi: 10.1101/2024.08.12.607489. bioRxiv. 2024. PMID: 39415996 Free PMC article. Preprint. - soibean: High-Resolution Taxonomic Identification of Ancient Environmental DNA Using Mitochondrial Pangenome Graphs.
Vogel NA, Rubin JD, Pedersen AG, Sackett PW, Pedersen MW, Renaud G. Vogel NA, et al. Mol Biol Evol. 2024 Oct 4;41(10):msae203. doi: 10.1093/molbev/msae203. Mol Biol Evol. 2024. PMID: 39361595 Free PMC article. - Deep-time phylogenetic inference by paleoproteomic analysis of dental enamel.
Taurozzi AJ, Rüther PL, Patramanis I, Koenig C, Sinclair Paterson R, Madupe PP, Harking FS, Welker F, Mackie M, Ramos-Madrigal J, Olsen JV, Cappellini E. Taurozzi AJ, et al. Nat Protoc. 2024 Jul;19(7):2085-2116. doi: 10.1038/s41596-024-00975-3. Epub 2024 Apr 26. Nat Protoc. 2024. PMID: 38671208 Review. - Benchmarking Metagenomic Classifiers on Simulated Ancient and Modern Metagenomic Data.
Pusadkar V, Azad RK. Pusadkar V, et al. Microorganisms. 2023 Oct 2;11(10):2478. doi: 10.3390/microorganisms11102478. Microorganisms. 2023. PMID: 37894136 Free PMC article. - Challenges and Opportunities behind the Use of Herbaria in Paleogenomics Studies.
Papalini S, Di Vittori V, Pieri A, Allegrezza M, Frascarelli G, Nanni L, Bitocchi E, Bellucci E, Gioia T, Pereira LG, Susek K, Tenaillon M, Neumann K, Papa R. Papalini S, et al. Plants (Basel). 2023 Sep 30;12(19):3452. doi: 10.3390/plants12193452. Plants (Basel). 2023. PMID: 37836192 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources