The case for cloud computing in genome informatics - PubMed (original) (raw)
The case for cloud computing in genome informatics
Lincoln D Stein. Genome Biol. 2010.
Abstract
With DNA sequencing now getting cheaper more quickly than data storage or computation, the time may have come for genome informatics to migrate to the cloud.
Figures
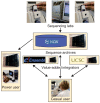
Figure 1
The old genome informatics ecosystem. Under the traditional flow of genome information, sequencing laboratories transmit raw and interpreted sequencing information across the internet to one of several sequencing archives. This information is accessed either directly by casual users or indirectly via a website run by one of the value-added genome integrators. Power users typically download large datasets from the archives onto their local compute clusters for computationally intensive number crunching. Under this model, the sequencing archives, value-added integrators and power users all maintain their own compute and storage clusters and keep local copies of the sequencing datasets.
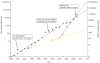
Figure 2
Historical trends in storage prices versus DNA sequencing costs. The blue squares describe the historic cost of disk prices in megabytes per US dollar. The long-term trend (blue line, which is a straight line here because the plot is logarithmic) shows exponential growth in storage per dollar with a doubling time of roughly 1.5 years. The cost of DNA sequencing, expressed in base pairs per dollar, is shown by the red triangles. It follows an exponential curve (yellow line) with a doubling time slightly slower than disk storage until 2004, when next generation sequencing (NGS) causes an inflection in the curve to a doubling time of less than 6 months (red line). These curves are not corrected for inflation or for the 'fully loaded' cost of sequencing and disk storage, which would include personnel costs, depreciation and overhead.
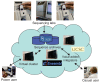
Figure 3
The 'new' genome informatics ecosystem based on cloud computing. In this model, the community's storage and compute resources are co-located in a 'cloud' maintained by a large service provider. The sequence archives and value-added integrators maintain servers and storage systems within the cloud, and use more or less capacity as needed for daily and seasonal fluctuations in usage. Casual users continue to access the data via the websites of the archives and integrators, but power users now have the option of creating virtual on-demand compute clusters within the cloud, which have direct access to the sequencing datasets.
Similar articles
- Gathering clouds and a sequencing storm: why cloud computing could broaden community access to next-generation sequencing.
[No authors listed] [No authors listed] Nat Biotechnol. 2010 Jan;28(1):1. doi: 10.1038/nbt0110-1. Nat Biotechnol. 2010. PMID: 20062015 No abstract available. - Businesses ready whole-genome analysis services for researchers.
Stokes T. Stokes T. Nat Med. 2011 Oct 11;17(10):1161. doi: 10.1038/nm1011-1161. Nat Med. 2011. PMID: 21988969 No abstract available. - Genome sequencing and assembly.
Grabherr MG, Mauceli E, Ma LJ. Grabherr MG, et al. Methods Mol Biol. 2011;722:1-9. doi: 10.1007/978-1-61779-040-9_1. Methods Mol Biol. 2011. PMID: 21590409 - Application of 'next-generation' sequencing technologies to microbial genetics.
MacLean D, Jones JD, Studholme DJ. MacLean D, et al. Nat Rev Microbiol. 2009 Apr;7(4):287-96. doi: 10.1038/nrmicro2122. Nat Rev Microbiol. 2009. PMID: 19287448 Review. - Computational solutions to large-scale data management and analysis.
Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Schadt EE, et al. Nat Rev Genet. 2010 Sep;11(9):647-57. doi: 10.1038/nrg2857. Nat Rev Genet. 2010. PMID: 20717155 Free PMC article. Review.
Cited by
- Collaborative cloud-enabled tools allow rapid, reproducible biological insights.
Ragan-Kelley B, Walters WA, McDonald D, Riley J, Granger BE, Gonzalez A, Knight R, Perez F, Caporaso JG. Ragan-Kelley B, et al. ISME J. 2013 Mar;7(3):461-4. doi: 10.1038/ismej.2012.123. Epub 2012 Oct 25. ISME J. 2013. PMID: 23096404 Free PMC article. No abstract available. - Recommendations on e-infrastructures for next-generation sequencing.
Spjuth O, Bongcam-Rudloff E, Dahlberg J, Dahlö M, Kallio A, Pireddu L, Vezzi F, Korpelainen E. Spjuth O, et al. Gigascience. 2016 Jun 7;5:26. doi: 10.1186/s13742-016-0132-7. Gigascience. 2016. PMID: 27267963 Free PMC article. Review. - Methods of integrating data to uncover genotype-phenotype interactions.
Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Ritchie MD, et al. Nat Rev Genet. 2015 Feb;16(2):85-97. doi: 10.1038/nrg3868. Epub 2015 Jan 13. Nat Rev Genet. 2015. PMID: 25582081 Review. - Novel bioinformatic developments for exome sequencing.
Lelieveld SH, Veltman JA, Gilissen C. Lelieveld SH, et al. Hum Genet. 2016 Jun;135(6):603-14. doi: 10.1007/s00439-016-1658-6. Epub 2016 Apr 13. Hum Genet. 2016. PMID: 27075447 Free PMC article. Review. - Adaptive efficient compression of genomes.
Wandelt S, Leser U. Wandelt S, et al. Algorithms Mol Biol. 2012 Nov 12;7(1):30. doi: 10.1186/1748-7188-7-30. Algorithms Mol Biol. 2012. PMID: 23146997 Free PMC article.
References
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources