Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus - PubMed (original) (raw)
Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus
Karen A O'Connell et al. J Virol. 2010 Jul.
Abstract
A subset of HIV-1-infected patients known as elite controllers or suppressors (ES) control the virus naturally. We have previously demonstrated sequence discordance between proviral and plasma gag clones in ES, much of which can be attributed to selective pressure from the host (J. R. Bailey, T. M. Williams, R. F. Siliciano, and J. N. Blankson, J. Exp. Med. 203:1357-1369, 2006). However, it is not clear whether ongoing viral replication continues in ES once the control of viremia has been established or whether selective pressure impacts this evolution. The cytotoxic T-lymphocyte (CTL) response in ES often targets Gag and frequently is superior to that of HIV-1 progressors, partially due to the HLA class I alleles B*57/5801 and B*27, which are overrepresented in ES. We therefore examined longitudinal plasma and proviral gag sequences from HLA-B*57/5801 and -B*27 ES. Despite the highly conserved nature of gag, we observed clear evidence of evolution in the plasma virus, largely due to synonymous substitutions. In contrast, evolution was rare in proviral clones, suggesting that ongoing replication in ES does not permit the significant reseeding of the latent reservoir. Interestingly, there was little continual evolution in CTL epitopes, and we detected de novo CTL responses to autologous viral mutants. Thus, some ES control viremia despite ongoing replication and evolution.
Figures
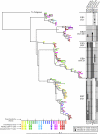
FIG. 1.
Phylogenetic analysis of gag in elite suppressor patients. Phylogenies were estimated by using a classical approach, functioning under a maximum-likelihood (ML) optimality criterion. All sequences are clonal, and APOBEC-mediated hypermutated sequences were removed from analysis. Bootstrap values of 50 and higher are displayed. Protective HLA B alleles are noted beneath the patient number. Patients are not related in any way but are placed on the same tree for display purposes. Colors indicate time, with the scale below in years. Triangles represent clonal plasma sequences, squares represent proviral sequences from HLA-DR+ CD4+ T cells, and circles represent clonal proviral sequences from HLA-DR− CD4+ T cells. To the right, escape mutations in HLA-B*57- and -B*27-restricted epitopes are denoted in black or gray and aligned with the appropriate symbol on the tree. Escape mutations are listed above the grid, with the epitopes in which the mutations occurred listed above the mutations.
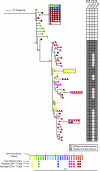
FIG. 2.
Phylogenetic analysis of gag in ES8. Phylogenies were constructed as described for Fig. 1; ES8 is isolated due to the large number of sequences obtained for this patient. Colors indicate time, with the scale below in years. Triangles represent plasma sequences, squares represent proviral sequences from HLA-DR+ CD4+ T cells, and circles represent proviral sequences from HLA-DR− CD4+ T cells. To the right, escape mutations in HLA B*57- and -B*27-restricted epitopes are denoted in black or gray and aligned with the appropriate symbol on the tree. Escape mutations are listed above the grid, with the epitopes in which the mutations occurred listed above the mutations.
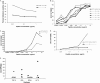
FIG. 3.
Analysis of synonymous and nonsynonymous mutation in the plasma virus and proviral compartments. Shown are p-distance values for plasma (A) and proviral (B) sequences as determined by comparing early and late samples for each patient utilizing the Nei-Gojobori method. The numbers of differences also were calculated for plasma (C) and proviral (D) sequences using the Nei-Gojobori method.
FIG. 4.
Relevant sequence regions from clonal, near-full-length gag amplified from plasma (pl) or from either resting (HLA-DR−) or activated (HLA-DR+) CD4+ T cells. The date of sample acquisition and number of clones that are identical to the displayed sequences are noted. The HLA-B*27-restricted epitope KK10 (Gag 263-272) and HLA-B*57-restricted epitopes IW9 (Gag 147-155), KF11 (Gag 162-172), TW10 (Gag 240-249), and QW9 (Gag 308-316) all are denoted in shaded boxes. Sites of compensatory mutations for KK10 (S173) (42) and TW10 (H219, I223, M228) escape mutants also are shaded, and for ES7 the DL15 epitope is displayed. Sequences from 2004 and 2005 for ES3, ES7, ES8, and ES9 have been previously reported and are shown for comparative purposes only.
FIG. 5.
(A) Intracellular IFN-γ staining analysis of ES1 HLA-B27 tetramer-positive cells to autologous and wild-type KK10 (KRWIILGLNK; Gag 263-272) peptides. (B) IFN-γ ELISPOT analysis of ES8 CD8+ T cells to autologous TW10 variants and wild-type TW10 peptide (TSTLQEQIGW; Gag 240-249). (C) IFN-γ ELISPOT analysis of ES31 CD8+ T cells to autologous TW10 variant and wild-type TW10 peptides. (D) IFN-γ ELISPOT analysis of the ES7 CD8+ T cells to the wild-type and the autologous peptide containing the K335R mutation in DL15 (DCKTILKALGPAATL; Gag 329-343). (E) Magnitude of IFN-γ response by ES8, ES3, ES7, and ES9 to four epitopes in which evolution occurred in the plasma virus of either ES7 or ES8. Data reflect the IFN-γ response to the autologous variant of the epitope of each patient at the latest time point available. Open symbols indicate SFC < 50. Circles indicate the response by patients in whom no evolution was seen in this epitope, while diamonds indicate the response by the patient in whom the evolution of the plasma virus occurred in this epitope (ES8 for TW10 and ES7 for IW9, QW9, and KF11).
Similar articles
- Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations.
Bailey JR, Williams TM, Siliciano RF, Blankson JN. Bailey JR, et al. J Exp Med. 2006 May 15;203(5):1357-69. doi: 10.1084/jem.20052319. Epub 2006 May 8. J Exp Med. 2006. PMID: 16682496 Free PMC article. - Evolution of the HIV-1 nef gene in HLA-B*57 positive elite suppressors.
Salgado M, Brennan TP, O'Connell KA, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Salgado M, et al. Retrovirology. 2010 Nov 8;7:94. doi: 10.1186/1742-4690-7-94. Retrovirology. 2010. PMID: 21059238 Free PMC article. - HLA-B*57 elite suppressor and chronic progressor HIV-1 isolates replicate vigorously and cause CD4+ T cell depletion in humanized BLT mice.
Salgado M, Swanson MD, Pohlmeyer CW, Buckheit RW 3rd, Wu J, Archin NM, Williams TM, Margolis DM, Siliciano RF, Garcia JV, Blankson JN. Salgado M, et al. J Virol. 2014 Mar;88(6):3340-52. doi: 10.1128/JVI.03380-13. Epub 2014 Jan 3. J Virol. 2014. PMID: 24390323 Free PMC article. - HIV-1 replicative fitness in elite controllers.
Lobritz MA, Lassen KG, Arts EJ. Lobritz MA, et al. Curr Opin HIV AIDS. 2011 May;6(3):214-20. doi: 10.1097/COH.0b013e3283454cf5. Curr Opin HIV AIDS. 2011. PMID: 21430530 Free PMC article. Review. - The implications of viral reservoirs on the elite control of HIV-1 infection.
Buckheit RW 3rd, Salgado M, Martins KO, Blankson JN. Buckheit RW 3rd, et al. Cell Mol Life Sci. 2013 Mar;70(6):1009-19. doi: 10.1007/s00018-012-1101-7. Epub 2012 Aug 4. Cell Mol Life Sci. 2013. PMID: 22864624 Free PMC article. Review.
Cited by
- Developing strategies for HIV-1 eradication.
Durand CM, Blankson JN, Siliciano RF. Durand CM, et al. Trends Immunol. 2012 Nov;33(11):554-62. doi: 10.1016/j.it.2012.07.001. Epub 2012 Aug 3. Trends Immunol. 2012. PMID: 22867874 Free PMC article. Review. - HIV controllers with different viral load cutoff levels have distinct virologic and immunologic profiles.
Côrtes FH, Passaes CP, Bello G, Teixeira SL, Vorsatz C, Babic D, Sharkey M, Grinsztejn B, Veloso V, Stevenson M, Morgado MG. Côrtes FH, et al. J Acquir Immune Defic Syndr. 2015 Apr 1;68(4):377-385. doi: 10.1097/QAI.0000000000000500. J Acquir Immune Defic Syndr. 2015. PMID: 25564106 Free PMC article. - Persistent HIV-1 replication during antiretroviral therapy.
Martinez-Picado J, Deeks SG. Martinez-Picado J, et al. Curr Opin HIV AIDS. 2016 Jul;11(4):417-23. doi: 10.1097/COH.0000000000000287. Curr Opin HIV AIDS. 2016. PMID: 27078619 Free PMC article. Review. - Primary CD8+ T cells from elite suppressors effectively eliminate non-productively HIV-1 infected resting and activated CD4+ T cells.
Buckheit RW 3rd, Siliciano RF, Blankson JN. Buckheit RW 3rd, et al. Retrovirology. 2013 Jul 1;10:68. doi: 10.1186/1742-4690-10-68. Retrovirology. 2013. PMID: 23816179 Free PMC article. - Long-term remission despite clonal expansion of replication-competent HIV-1 isolates.
Veenhuis RT, Kwaa AK, Garliss CC, Latanich R, Salgado M, Pohlmeyer CW, Nobles CL, Gregg J, Scully EP, Bailey JR, Bushman FD, Blankson JN. Veenhuis RT, et al. JCI Insight. 2018 Sep 20;3(18):e122795. doi: 10.1172/jci.insight.122795. eCollection 2018 Sep 20. JCI Insight. 2018. PMID: 30232278 Free PMC article.
References
- Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473-2485. - PMC - PubMed
- Andrade, A., J. R. Bailey, J. Xu, F. H. Philp, T. C. Quinn, T. M. Williams, S. C. Ray, D. L. Thomas, and J. N. Blankson. 2008. CD4+ T-cell depletion in an untreated HIV type 1-infected human leukocyte antigen-B*5801-positive patient with an undetectable viral load. Clin. Infect. Dis. 46:e78-e82. - PMC - PubMed
- Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441-6457. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials