Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae - PubMed (original) (raw)
Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae
Jiefei Geng et al. Mol Biol Cell. 2010.
Abstract
In eukaryotic cells, autophagy mediates the degradation of cytosolic contents in response to environmental change. Genetic analyses in fungi have identified over 30 autophagy-related (ATG) genes and provide substantial insight into the molecular mechanism of this process. However, one essential issue that has not been resolved is the origin of the lipids that form the autophagosome, the sequestering vesicle that is critical for autophagy. Here, we report that two post-Golgi proteins, Sec2 and Sec4, are required for autophagy. Sec4 is a Rab family GTPase, and Sec2 is its guanine nucleotide exchange factor. In sec2 and sec4 conditional mutant yeast, the anterograde movement of Atg9, a proposed membrane carrier, is impaired during starvation conditions. Similarly, in the sec2 mutant, Atg8 is inefficiently recruited to the phagophore assembly site, which is involved in autophagosome biogenesis, resulting in the generation of fewer autophagosomes. We propose that following autophagy induction the function of Sec2 and Sec4 are diverted to direct membrane flow to autophagosome formation.
Figures
Figure 1.
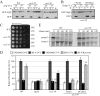
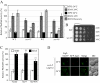
Sec2 is involved in both autophagy and the Cvt pathway. (A) GFP-Atg8 processing is blocked in sec2 mutants. GFP-Atg8 was expressed by chromosomal integration in wild-type (JGY146), sec2-59 (JGY183), and sec2-41 (JGY184) strains. Cells were cultured in YPD at 24°C to midlog phase. For each strain, the culture was divided into two parts. One-half was incubated at 37°C for 30 min to inactivate the sec2 mutant, whereas the other one remained at 24°C. Then cells were shifted to SD-N and incubated for 2 h at the same temperature. Samples were taken before and after starvation. For recovery (R), cells starved at 37°C for 2 h were shifted back to 24°C and starved for another 2 h. Immunoblotting was done with anti-YFP antibody (that recognizes GFP) and the positions of full-length GFP-Atg8 and free GFP are indicated. (B) Exogenous expression of Sec2 restores autophagy in the sec2-59 mutant. sec2-59 GFP-Atg8 cells (JGY183) harboring an empty vector or pCuSec2(416) were cultured in SMD medium and examined as described in A. (C) sec2 mutants are viable after 2-h starvation at 37°C. The same amount of starved cells from A were diluted and plated on a YPD plate, followed by 2-d incubation at room temperature. Cells were diluted 1:5 in each step from left to right. (D) Pho8Δ60 activity in the sec2-59 mutant. Wild-type (TN124), _atg1_Δ (HAY572), and sec2-59 (JGY112) cells as well as sec2-59 cells expressing an empty vector or pCuSec2(416) were cultured as in A. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bar, SD of three independent experiments. Significant difference compared with wild-type or vector alone, **p < 0.01. (E) The Cvt pathway is defective in the sec2-59 mutant. Wild-type (SEY6210) and sec2-59 (JGY113) cells were grown in SMD at 24°C. After a 30-min inactivation at 37°C, cells were subjected to pulse-chase labeling as described in Materials and Methods.
Figure 2.
A 58-amino acid domain on Sec2 is essential for its role in autophagy. (A) Schematic representation of Sec2 alleles. (B) Mutations within the 58-amino acid domain result in defective GFP-Atg8 processing. Wild-type (JGY124), sec2-78 (JGY125) and _sec2(Δ_451-508) (JGY126) cells were transformed with the pGFP-Atg8(316) plasmid and examined as in Figure 1A. (C) An autophagic defect in sec2-78 and _sec2(Δ_451-508) is confirmed by the Pho8Δ60 assay. The corresponding Pho8Δ60 strains (JGY127, JGY128, and JGY129) to those used in B were examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with wild-type, **p < 0.01. (D) The extreme C terminus of Sec2 is dispensable to complement the _sec2-59_ mutant. _sec2-59_ (JGY112) cells were transformed with plasmids expressing various truncated forms of Sec2 and examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with vector alone, **p < 0.01; no significant difference, ○p > 0.1.
Figure 3.
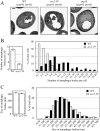
Sec2 mutation results in a reduced number of autophagic bodies, but has no effect on their size. After inactivation, the wild-type (_pep4_Δ _vps4_Δ, FRY143), _atg1_Δ (_atg1_Δ _pep4_Δ _vps4_Δ, HCY76), and sec2-59 (_sec2-59 pep4_Δ _vps4_Δ, JGY180) strains were starved for 2 h at 37°C and then analyzed by EM as described in Materials and Methods. (A) Representative images of wild-type, _atg1_Δ, and sec2-59 strains. Scale bar, 500 nm. (B and C) Number and size of autophagic bodies. Quantification was done as described in Materials and Methods. Error bars, SEM.
Figure 4.
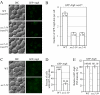
Recruitment of Atg8 to the PAS is affected in the sec2-59 mutant. (A and B) In the vam3ts background, sec2-59 cells accumulate fewer GFP-Atg8 dots in the cytosol. After preinactivation, vam3ts (JGY169) and vam3ts sec2 mutants (JGY171, JGY172) expressing chromosomally integrated GFP-Atg8 were starved for 20 min at 37°C. Cells were fixed to avoid any movement of GFP-Atg8, and fluorescence microscopy was performed as described in Materials and Methods. Twelve Z-section images were projected to visualize all dots present throughout the entire cell. Representative images are shown in A, and quantification of the number of GFP-Atg8 puncta is shown in B. Error bar, SEM; n = 100. (C–E) PAS intensity of GFP-Atg8 is not affected in the sec2-59 mutant although fewer cells have GFP-Atg8 dots. The wild-type (JGY146) and sec2-59 (JGY183) GFP-Atg8 strains were incubated in SD-N for 20 min at 37°C, and cells were examined by microscopy. (C) Representative images after projection of 12 Z-section pictures. (D) The percentage of cells that had at least one GFP-Atg8 dot was determined. Error bar, SD from three independent experiments; n = 300. (E) Relative PAS intensity of GFP-Atg8. The intensity in wild-type cells was set to 100%, and the other value was normalized. Error bar, SEM; n = 50. Scale bar, 5 μm.
Figure 5.
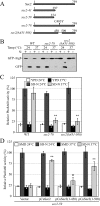
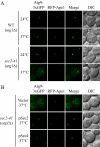
Atg9 anterograde movement is less efficient in the sec2 mutant. (A) In the TAKA assay, Atg9 is localized to multiple dots in the sec2-41 mutant at the NPT. Wild-type (Atg9-3xGFP RFP-Ape1 _atg1_Δ, JGY197) and sec2-41 mutant (Atg9-3xGFP RFP-Ape1 _atg1_Δ, JGY107) cells were starved at the indicated temperature for 2 h. Cells were fixed and then analyzed by microscopy. 12 Z-section images were stacked, and representative images are shown. (B) Exogenous expression of Sec4 can restore the Atg9 anterograde movement defect in sec2-41 cells as shown by the TAKA assay. sec2-41 (Atg9-3xGFP RFP-Ape1 _atg1_Δ) cells were transformed with empty vector, pSec2(413) or pSec4(413). Microscopy samples were prepared, and pictures were taken as described in A. Scale bar, 2 μm.
Figure 6.
The sec4 mutant displays an autophagic defect. (A) Two sec4 alleles show a defect in Pho8Δ60 activity. The sec4-2 (JGY168) and sec4-8 (JGY159) mutants and the corresponding wild-type (JGY166, YTS158) strains were cultured in YPD medium at 24°C to midlog phase. Half of the culture was inactivated for Sec4 function at 34°C for 30 min, whereas the other half was kept growing at 24°C. Then cells were shifted to SD-N, and the temperature was maintained. Samples were collected before and after 2-h starvation. For recovery, cells starved at 34°C were shifted back to 24°C and incubated for another 2 h. Error bar, SD from three independent assays. Significant difference compared with corresponding wild-type cells, **p < 0.01. (B) The _sec4_ mutant is viable after 2-h starvation at 34°C. The same amount of wild-type (YTS158) and _sec4-8_ (JGY159) cells after 2-h starvation at the indicated temperature were diluted and spotted on a YPD plate. Cells were diluted 1:5 in each step from left to right. The plate was incubated at room temperature for 2 d. (C) Sec4S34N has a dominant-negative effect on autophagy. Wild-type cells (TN124) were transformed with an empty vector or a plasmid expressing either Sec4 or Sec4S34N under the control of the _CUP1_ promoter. Transformants were cultured in SMD medium at 30°C and shifted to SD-N for 2 h. Before and after starvation, samples were collected and tested by the Pho8Δ60 assay. Error bar, SD from three independent assays. Significant difference compared with vector alone, **p < 0.01; no significant difference, ○p > 0.1. (D) Atg9 movement to the PAS is defective in the sec4-8 mutant. sec4-8 (Atg9-3xGFP RFP-Ape1 _atg1_Δ, JGY198) cells were used for microscopy and the procedure was similar to that described in Figure 5A. Scale bar, 2 μm.
Figure 7.
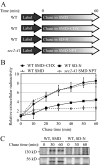
Protein secretion is down-regulated during starvation conditions. (A) Schematic representation of the procedure used for the pulse-chase experiments. (B) Relative radioactivity of the extracellular media. Cells were chased in the indicated media at 30°C, except for the sec2-41 cells, which were chased at 37°C. Samples were collected at the indicated time points. Extracellular proteins were precipitated and the amount of radioactivity quantified using a scintillation counter. The values at time zero were set to 1.0, and other values were normalized. Error bar, SD from three independent experiments. (C) Secretion of extracellular protein was examined by SDS-PAGE. Samples collected at 0, 30, and 60 min from B were separated by SDS-PAGE and visualized by autoradiography.
Figure 8.
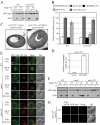
Autophagy defect and abnormal Atg8 localization in the ypt31/32ts mutant. (A) GFP-Atg8 processing is blocked in the ypt31/32ts mutant as shown by GFP-Atg8 processing. Wild-type (NSY128) and ypt31/32ts (NSY340) cells transformed with the pGFP-Atg8(316) plasmid were grown in SMD at 24°C. Samples were collected after incubation at 24 or 37°C and examined as described in Figure 1A. Recovery (R) refers to cells shifted back to 24°C after the 37°C incubation. (B) Pho8Δ60 assay in the ypt31/32ts mutant. Wild-type (JGY220) and ypt31/32ts (JGY209) cells were examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with wild-type, **p < 0.01. (C and D) On starvation, no autophagic bodies were accumulated in ypt31/32ts cells. _ypt31/32ts pep4_Δ _vps4_Δ (JGY222) cells harboring pCu416 or pCuYpt31(416) were cultured in SMD at 24°C. After inactivation for 30 min, cells were starved for 2 h at the NPT. EM samples were prepared, and EM data were analyzed as described in Figure 3. (C) Representative EM images. Scale bar, 500 nm. The number of autophagic bodies was quantified and shown in D. Error bars, SEM. (E and F) ypt31/32ts is defective in Atg8 localization. Wild-type (JGY217), vam3ts (JGY227), and ypt31/32ts (JGY210) cells were cultured in YPD medium at 24°C, shifted to 37°C for 30 min, and subsequently starved at 37°C. To recover the temperature-sensitive phenotype, vam3ts and ypt31/32ts cells were shifted back to 24°C and incubated in YPD or SD-N medium for another 2 h. (E) Representative fluorescence microscopy images. The vacuolar membrane was stained with FM 4-64 as described in Materials and Methods. For each condition, only one Z-section with a clearly defined vacuole was shown. Scale bar, 2 μm. (F) Samples taken at the indicated conditions were examined by immunoblotting using anti-YFP antibody. (G) Ypt31/32 are not involved in movement of Atg9 toward the PAS. ypt31/32ts (Atg9-3xGFP RFP-Ape1 _atg1_Δ, JGY192) mutant cells were examined using the TAKA assay and the procedure was similar to that described in Figure 5A. Scale bar, 2 μm.
Similar articles
- Activation of Rab GTPase Sec4 by its GEF Sec2 is required for prospore membrane formation during sporulation in yeast Saccharomyces cerevisiae.
Suda Y, Tachikawa H, Inoue I, Kurita T, Saito C, Kurokawa K, Nakano A, Irie K. Suda Y, et al. FEMS Yeast Res. 2018 Feb 1;18(1). doi: 10.1093/femsyr/fox095. FEMS Yeast Res. 2018. PMID: 29293994 - Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae.
van der Vaart A, Griffith J, Reggiori F. van der Vaart A, et al. Mol Biol Cell. 2010 Jul 1;21(13):2270-84. doi: 10.1091/mbc.e09-04-0345. Epub 2010 May 5. Mol Biol Cell. 2010. PMID: 20444982 Free PMC article. - The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.
Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. Yen WL, et al. J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article. - Regulation of membrane traffic by Rab GEF and GAP cascades.
Novick P. Novick P. Small GTPases. 2016 Oct;7(4):252-256. doi: 10.1080/21541248.2016.1213781. Epub 2016 Jul 18. Small GTPases. 2016. PMID: 27427966 Free PMC article. Review. - Mechanisms of autophagosome biogenesis.
Rubinsztein DC, Shpilka T, Elazar Z. Rubinsztein DC, et al. Curr Biol. 2012 Jan 10;22(1):R29-34. doi: 10.1016/j.cub.2011.11.034. Curr Biol. 2012. PMID: 22240478 Review.
Cited by
- Phosphatidylinositol 4-kinases are required for autophagic membrane trafficking.
Wang K, Yang Z, Liu X, Mao K, Nair U, Klionsky DJ. Wang K, et al. J Biol Chem. 2012 Nov 2;287(45):37964-72. doi: 10.1074/jbc.M112.371591. Epub 2012 Sep 13. J Biol Chem. 2012. PMID: 22977244 Free PMC article. - Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures.
Kaiser SE, Mao K, Taherbhoy AM, Yu S, Olszewski JL, Duda DM, Kurinov I, Deng A, Fenn TD, Klionsky DJ, Schulman BA. Kaiser SE, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1242-9. doi: 10.1038/nsmb.2415. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142976 Free PMC article. - Autophagy in Tenebrio molitor Immunity: Conserved Antimicrobial Functions in Insect Defenses.
Jo YH, Lee JH, Patnaik BB, Keshavarz M, Lee YS, Han YS. Jo YH, et al. Front Immunol. 2021 May 31;12:667664. doi: 10.3389/fimmu.2021.667664. eCollection 2021. Front Immunol. 2021. PMID: 34135896 Free PMC article. Review. - Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation.
Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Romanov J, et al. EMBO J. 2012 Nov 14;31(22):4304-17. doi: 10.1038/emboj.2012.278. Epub 2012 Oct 12. EMBO J. 2012. PMID: 23064152 Free PMC article. - Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy.
Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang HG. Takahashi Y, et al. Autophagy. 2011 Jan;7(1):61-73. doi: 10.4161/auto.7.1.14015. Epub 2011 Jan 1. Autophagy. 2011. PMID: 21068542 Free PMC article.
References
- Babu P., Bryan J. D., Panek H. R., Jordan S. L., Forbrich B. M., Kelley S. C., Colvin R. T., Robinson L. C. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 2002;115:4957–4968. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases