A draft sequence of the Neandertal genome - PubMed (original) (raw)
Comparative Study
. 2010 May 7;328(5979):710-722.
doi: 10.1126/science.1188021.
Johannes Krause # 1, Adrian W Briggs # 1, Tomislav Maricic # 1, Udo Stenzel # 1, Martin Kircher # 1, Nick Patterson # 2, Heng Li 2, Weiwei Zhai 3, Markus Hsi-Yang Fritz 4, Nancy F Hansen 5, Eric Y Durand 3, Anna-Sapfo Malaspinas 3, Jeffrey D Jensen 6, Tomas Marques-Bonet 7 8, Can Alkan 7, Kay Prüfer 1, Matthias Meyer 1, Hernán A Burbano 1, Jeffrey M Good 1 9, Rigo Schultz 1, Ayinuer Aximu-Petri 1, Anne Butthof 1, Barbara Höber 1, Barbara Höffner 1, Madlen Siegemund 1, Antje Weihmann 1, Chad Nusbaum 2, Eric S Lander 2, Carsten Russ 2, Nathaniel Novod 2, Jason Affourtit 10, Michael Egholm 10, Christine Verna 11, Pavao Rudan 12, Dejana Brajkovic 13, Željko Kucan 12, Ivan Gušic 12, Vladimir B Doronichev 14, Liubov V Golovanova 14, Carles Lalueza-Fox 8, Marco de la Rasilla 15, Javier Fortea 15, Antonio Rosas 16, Ralf W Schmitz 17 18, Philip L F Johnson 19, Evan E Eichler 7, Daniel Falush 20, Ewan Birney 4, James C Mullikin 5, Montgomery Slatkin 3, Rasmus Nielsen 3, Janet Kelso 1, Michael Lachmann 1, David Reich 2 21, Svante Pääbo 1
Affiliations
- PMID: 20448178
- PMCID: PMC5100745
- DOI: 10.1126/science.1188021
Comparative Study
A draft sequence of the Neandertal genome
Richard E Green et al. Science. 2010.
Abstract
Neandertals, the closest evolutionary relatives of present-day humans, lived in large parts of Europe and western Asia before disappearing 30,000 years ago. We present a draft sequence of the Neandertal genome composed of more than 4 billion nucleotides from three individuals. Comparisons of the Neandertal genome to the genomes of five present-day humans from different parts of the world identify a number of genomic regions that may have been affected by positive selection in ancestral modern humans, including genes involved in metabolism and in cognitive and skeletal development. We show that Neandertals shared more genetic variants with present-day humans in Eurasia than with present-day humans in sub-Saharan Africa, suggesting that gene flow from Neandertals into the ancestors of non-Africans occurred before the divergence of Eurasian groups from each other.
Figures
Fig. 1
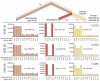
Samples and sites from which DNA was retrieved. (A) The three bones from Vindija from which Neandertal DNA was sequenced. (B) Map showing the four archaeological sites from which bones were used and their approximate dates (years B.P.).
Fig. 2
Nucleotide substitutions inferred to have occurred on the evolutionary lineages leading to the Neandertals, the human, and the chimpanzee genomes. In red are substitutions on the Neandertal lineage, in yellow the human lineage, and in pink the combined lineage from the common ancestor of these to the chimpanzee. For each lineage and each bone from Vindija, the distributions and numbers of substitutions are shown. The excess of C to T and G to A substitutions are due to deamination of cytosine residues in the Neandertal DNA.
Fig. 3
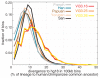
Divergence of Neandertal and human genomes. Distributions of divergence from the human genome reference sequence among segments of 100 kb are shown for three Neandertals and the five present-day humans.
Fig. 4
Selective sweep screen. (A) Schematic illustration of the rationale for the selective sweep screen. For many regions of the genome, the variation within current humans is old enough to include Neandertals (left). Thus, for SNPs in present-day humans, Neandertals often carry the derived allele (blue). However, in genomic regions where an advantageous mutation arises (right, red star) and sweeps to high frequency or fixation in present-day humans, Neandertals will be devoid of derived alleles. (B) Candidate regions of selective sweeps. All 4235 regions of at least 25 kb where S (see SOM Text 13) falls below two standard deviations of the mean are plotted by their S and genetic width. Regions on the autosomes are shown in orange and those on the X chromosome in blue. The top 5% by S are shadowed in light blue. (C) The top candidate region from the selective sweep screen contains two genes, ZFP36L2 and THADA. The red line shows the log-ratio of the number of observed Neandertal-derived alleles versus the number of expected Neandertal-derived alleles, within a 100 kilobase window. The blue dots above the panel indicate all SNP positions, and the green dots indicate SNPs where the Neandertal carries the derived allele.
Fig. 5
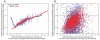
Segments of Neandertal ancestry in the human reference genome. We examined 2825 segments in the human reference genome that are of African ancestry and 2797 that are of European ancestry. (A) European segments, with few differences from the Neandertals, tend to have many differences from other present-day humans, whereas African segments do not, as expected if the former are derived from Neandertals. (B) Scatter plot of the segments in (A) with respect to their divergence to the Neandertals and to Venter. In the top left quandrant, 94% of segments are of European ancestry, suggesting that many of them are due to gene flow from Neandertals.
Fig. 6
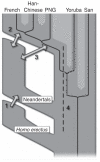
Four possible scenarios of genetic mixture involving Neandertals. Scenario 1 represents gene flow into Neandertal from other archaic hominins, here collectively referred to as Homo erectus. This would manifest itself as segments of the Neandertal genome with unexpectedly high divergence from present-day humans. Scenario 2 represents gene flow between late Neandertals and early modern humans in Europe and/or western Asia. We see no evidence of this because Neandertals are equally distantly related to all non-Africans. However, such gene flow may have taken place without leaving traces in the present-day gene pool. Scenario 3 represents gene flow between Neandertals and the ancestors of all non-Africans. This is the most parsimonious explanation of our observation. Although we detect gene flow only from Neandertals into modern humans, gene flow in the reverse direction may also have occurred. Scenario 4 represents old substructure in Africa that persisted from the origin of Neandertals until the ancestors of non-Africans left Africa. This scenario is also compatible with the current data.
Comment in
- Paleogenetics. Close encounters of the prehistoric kind.
Gibbons A. Gibbons A. Science. 2010 May 7;328(5979):680-4. doi: 10.1126/science.328.5979.680. Science. 2010. PMID: 20448163 No abstract available. - Genomics: Technical feat gives clues to human origins.
Casci T. Casci T. Nat Rev Genet. 2010 Jul;11(7):456. doi: 10.1038/nrg2821. Nat Rev Genet. 2010. PMID: 20517343 No abstract available.
Similar articles
- Paleogenetics. Close encounters of the prehistoric kind.
Gibbons A. Gibbons A. Science. 2010 May 7;328(5979):680-4. doi: 10.1126/science.328.5979.680. Science. 2010. PMID: 20448163 No abstract available. - IBD Sharing between Africans, Neandertals, and Denisovans.
Povysil G, Hochreiter S. Povysil G, et al. Genome Biol Evol. 2016 Dec 1;8(12):3406-3416. doi: 10.1093/gbe/evw234. Genome Biol Evol. 2016. PMID: 28158547 Free PMC article. - Targeted investigation of the Neandertal genome by array-based sequence capture.
Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, Meyer M, Good JM, Maricic T, Johnson PL, Xuan Z, Rooks M, Bhattacharjee A, Brizuela L, Albert FW, de la Rasilla M, Fortea J, Rosas A, Lachmann M, Hannon GJ, Pääbo S. Burbano HA, et al. Science. 2010 May 7;328(5979):723-5. doi: 10.1126/science.1188046. Science. 2010. PMID: 20448179 Free PMC article. - The Neandertal genome and ancient DNA authenticity.
Green RE, Briggs AW, Krause J, Prüfer K, Burbano HA, Siebauer M, Lachmann M, Pääbo S. Green RE, et al. EMBO J. 2009 Sep 2;28(17):2494-502. doi: 10.1038/emboj.2009.222. Epub 2009 Aug 6. EMBO J. 2009. PMID: 19661919 Free PMC article. Review. - [Progresses on Neandertal genomics].
Bi CL, Guo GY, Zhang X, Tian YH, Shen YZ. Bi CL, et al. Yi Chuan. 2012 Jun;34(6):659-65. doi: 10.3724/sp.j.1005.2012.00659. Yi Chuan. 2012. PMID: 22698735 Review. Chinese.
Cited by
- The Combined Landscape of Denisovan and Neanderthal Ancestry in Present-Day Humans.
Sankararaman S, Mallick S, Patterson N, Reich D. Sankararaman S, et al. Curr Biol. 2016 May 9;26(9):1241-7. doi: 10.1016/j.cub.2016.03.037. Epub 2016 Mar 28. Curr Biol. 2016. PMID: 27032491 Free PMC article. - Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers.
Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo JM, Lema G, Fu W, Nyambo TB, Rebbeck TR, Zhang K, Akey JM, Tishkoff SA. Lachance J, et al. Cell. 2012 Aug 3;150(3):457-69. doi: 10.1016/j.cell.2012.07.009. Epub 2012 Jul 26. Cell. 2012. PMID: 22840920 Free PMC article. - Multiple Horizontal Mini-chromosome Transfers Drive Genome Evolution of Clonal Blast Fungus Lineages.
Barragan AC, Latorre SM, Malmgren A, Harant A, Win J, Sugihara Y, Burbano HA, Kamoun S, Langner T. Barragan AC, et al. Mol Biol Evol. 2024 Aug 2;41(8):msae164. doi: 10.1093/molbev/msae164. Mol Biol Evol. 2024. PMID: 39107250 Free PMC article. - Multiple Genomic Events Altering Hominin SIGLEC Biology and Innate Immunity Predated the Common Ancestor of Humans and Archaic Hominins.
Khan N, de Manuel M, Peyregne S, Do R, Prufer K, Marques-Bonet T, Varki N, Gagneux P, Varki A. Khan N, et al. Genome Biol Evol. 2020 Jul 1;12(7):1040-1050. doi: 10.1093/gbe/evaa125. Genome Biol Evol. 2020. PMID: 32556248 Free PMC article. - Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa.
Campbell MC, Ranciaro A, Froment A, Hirbo J, Omar S, Bodo JM, Nyambo T, Lema G, Zinshteyn D, Drayna D, Breslin PA, Tishkoff SA. Campbell MC, et al. Mol Biol Evol. 2012 Apr;29(4):1141-53. doi: 10.1093/molbev/msr293. Epub 2011 Nov 29. Mol Biol Evol. 2012. PMID: 22130969 Free PMC article.
References
- Bischoff JL, et al. High-Resolution U-Series Dates from the Sima de los Huesos Hominids Yields 600+/−66 kyrs: Implications for the Evolution of the Early Neanderthal Lineage. Vol. 34. Elsevier; Amsterdam, PAYS-BAS: 2007.
- Stringer CB, Hublin J. J. Hum. Evol. 1999;37:873. - PubMed
- Finlayson C, et al. Nature. 2006;443:850. - PubMed
- Krause J, et al. Nature. 2007;449:902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM040282/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- ImNIH/Intramural NIH HHS/United States
- GM40282/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials