Regulation and isoform function of the V-ATPases - PubMed (original) (raw)
Review
Regulation and isoform function of the V-ATPases
Masashi Toei et al. Biochemistry. 2010.
Abstract
The vacuolar (H(+))-ATPases are ATP-dependent proton pumps that acidify intracellular compartments and, in some cases, transport protons across the plasma membrane of eukaryotic cells. Intracellular V-ATPases play an important role in normal physiological processes such as receptor-mediated endocytosis, intracellular membrane trafficking, pro-hormone processing, protein degradation, and the coupled uptake of small molecules, such as neurotransmitters. They also function in the entry of various pathogenic agents, including many envelope viruses, like influenza virus, and toxins, like anthrax toxin. Plasma membrane V-ATPases function in renal pH homeostasis, bone resorption and sperm maturation, and various disease processes, including renal tubular acidosis, osteopetrosis, and tumor metastasis. V-ATPases are composed of a peripheral V(1) domain containing eight different subunits that is responsible for ATP hydrolysis and an integral V(0) domain containing six different subunits that translocates protons. In mammalian cells, most of the V-ATPase subunits exist in multiple isoforms which are often expressed in a tissue specific manner. Isoforms of one of the V(0) subunits (subunit a) have been shown to possess information that targets the V-ATPase to distinct cellular destinations. Mutations in isoforms of subunit a lead to the human diseases osteopetrosis and renal tubular acidosis. A number of mechanisms are employed to regulate V-ATPase activity in vivo, including reversible dissociation of the V(1) and V(0) domains, control of the tightness of coupling of proton transport and ATP hydrolysis, and selective targeting of V-ATPases to distinct cellular membranes. Isoforms of subunit a are involved in regulation both via the control of coupling and via selective targeting. This review will begin with a brief introduction to the function, structure, and mechanism of the V-ATPases followed by a discussion of the role of V-ATPase subunit isoforms and the mechanisms involved in regulation of V-ATPase activity.
Figures
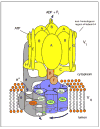
Fig. 1. Structure and mechanism of the V-ATPase
The V-ATPase is composed of two domains, V1 and V0. The peripheral V1 domain is composed of eight different subunits (A-H, shown in yellow and orange) and is responsible for ATP hydrolysis, whereas the integral V0 domain is composed of six subunits (in yeast these are subunits a, c, c′, c″, d and e, shown in blue and grey), and is involved in proton translocation across the membrane. ATP hydrolysis drives the rotation of a central rotor, which is composed of the D, F, d and proteolipid (c, c′, c″) subunits. Subunit a possesses two hemi-channels and a crucial arginine residue (shown in red), which are required for proton translocation. The hemi-channels allow protons to reach buried glutamic acid residues (shown in green) on the proteolipid ring from the cytoplasmic side of the membrane and to leave from these sites to the luminal side of the membrane following interaction of the glutamate residues with the a subunit arginine residue. The V1 and V0 domain are connected by a central stalk, which is composed of subunits D, F and d, and three peripheral stalks, which are composed of subunits C, E, G and the N-terminal cytoplasmic domain of subunit a. The peripheral stalks hold the A3B3 hexamer stationary with respect to subunit a. The non-homologous region of subunit A, which is absent from the F1FO ATP synthases, is involved in reversible dissociation (see text).
Fig. 2. Regulation of V-ATPase activity by reversible dissociation
In yeast, glucose depletion initiates the reversible dissociation of the V-ATPase into a V1 complex minus C, free subunit C and V0. Dissociation requires an intact microtubule network whereas reassembly of V1 back onto V0 is mediated by the RAVE complex (composed of Skp1, Rav1 and Rav2) and aldolase. Dissociation may also be stimulated by altered interactions of the non-homologous region of subunit A with the V0 domain. Glucose-dependent assembly of the V-ATPase is driven by activation of the Ras/cAMP/protein kinase A (PKA) pathway. The free forms of V1 and V0 are silenced with respect to ATP hydrolysis and passive proton translocation.
Similar articles
- Function, structure and regulation of the vacuolar (H+)-ATPases.
Jefferies KC, Cipriano DJ, Forgac M. Jefferies KC, et al. Arch Biochem Biophys. 2008 Aug 1;476(1):33-42. doi: 10.1016/j.abb.2008.03.025. Epub 2008 Mar 29. Arch Biochem Biophys. 2008. PMID: 18406336 Free PMC article. Review. - Structure and regulation of the vacuolar ATPases.
Cipriano DJ, Wang Y, Bond S, Hinton A, Jefferies KC, Qi J, Forgac M. Cipriano DJ, et al. Biochim Biophys Acta. 2008 Jul-Aug;1777(7-8):599-604. doi: 10.1016/j.bbabio.2008.03.013. Epub 2008 Mar 29. Biochim Biophys Acta. 2008. PMID: 18423392 Free PMC article. - Subunit structure, function, and arrangement in the yeast and coated vesicle V-ATPases.
Inoue T, Wilkens S, Forgac M. Inoue T, et al. J Bioenerg Biomembr. 2003 Aug;35(4):291-9. doi: 10.1023/a:1025720713747. J Bioenerg Biomembr. 2003. PMID: 14635775 Review. - Structure, mechanism and regulation of the clathrin-coated vesicle and yeast vacuolar H(+)-ATPases.
Forgac M. Forgac M. J Exp Biol. 2000 Jan;203(Pt 1):71-80. doi: 10.1242/jeb.203.1.71. J Exp Biol. 2000. PMID: 10600675 - V-ATPase functions in normal and disease processes.
Hinton A, Bond S, Forgac M. Hinton A, et al. Pflugers Arch. 2009 Jan;457(3):589-98. doi: 10.1007/s00424-007-0382-4. Epub 2007 Nov 20. Pflugers Arch. 2009. PMID: 18026982 Review.
Cited by
- Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1.
Zhang T, Zhou Q, Ogmundsdottir MH, Möller K, Siddaway R, Larue L, Hsing M, Kong SW, Goding CR, Palsson A, Steingrimsson E, Pignoni F. Zhang T, et al. J Cell Sci. 2015 Aug 1;128(15):2938-50. doi: 10.1242/jcs.173807. Epub 2015 Jun 19. J Cell Sci. 2015. PMID: 26092939 Free PMC article. - Leishmania amazonensis sabotages host cell SUMOylation for intracellular survival.
Okuda K, Silva Costa Franco MM, Yasunaga A, Gazzinelli R, Rabinovitch M, Cherry S, Silverman N. Okuda K, et al. iScience. 2022 Aug 13;25(9):104909. doi: 10.1016/j.isci.2022.104909. eCollection 2022 Sep 16. iScience. 2022. PMID: 36060064 Free PMC article. - β-Catenin-dependent lysosomal targeting of internalized tumor necrosis factor-α suppresses caspase-8 activation in apoptosis-resistant colon cancer cells.
Han J, Sridevi P, Ramirez M, Ludwig KJ, Wang JY. Han J, et al. Mol Biol Cell. 2013 Feb;24(4):465-73. doi: 10.1091/mbc.E12-09-0662. Epub 2012 Dec 21. Mol Biol Cell. 2013. PMID: 23264463 Free PMC article. - Osteopetrosis mutation R444L causes endoplasmic reticulum retention and misprocessing of vacuolar H+-ATPase a3 subunit.
Bhargava A, Voronov I, Wang Y, Glogauer M, Kartner N, Manolson MF. Bhargava A, et al. J Biol Chem. 2012 Aug 3;287(32):26829-39. doi: 10.1074/jbc.M112.345702. Epub 2012 Jun 8. J Biol Chem. 2012. PMID: 22685294 Free PMC article. - Hijacking the T-cell communication network by the human T-cell leukemia/lymphoma virus type 1 (HTLV-1) p12 and p8 proteins.
Van Prooyen N, Andresen V, Gold H, Bialuk I, Pise-Masison C, Franchini G. Van Prooyen N, et al. Mol Aspects Med. 2010 Oct;31(5):333-43. doi: 10.1016/j.mam.2010.07.001. Epub 2010 Jul 29. Mol Aspects Med. 2010. PMID: 20673780 Free PMC article. Review.
References
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. - PubMed
- Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. - PubMed
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. - PubMed
- Gu F, Gruenberg J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J Biol Chem. 2000;275:8154–8160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM034478/GM/NIGMS NIH HHS/United States
- R37 GM034478-26/GM/NIGMS NIH HHS/United States
- GM34478/GM/NIGMS NIH HHS/United States
- R37 GM034478/GM/NIGMS NIH HHS/United States
- R37 GM034478-25/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases