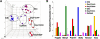Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis - PubMed (original) (raw)
Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis
Fuchou Tang et al. Cell Stem Cell. 2010.
Abstract
During the transition from the inner cell mass (ICM) cells of blastocysts to pluripotent embryonic stem cells (ESCs) in vitro, a normal developmental program is replaced in cells that acquire a capacity for infinite self-renewal and pluripotency. We explored the underlying mechanism of this switch by using RNA-Seq transcriptome analysis at the resolution of single cells. We detected significant molecular transitions and major changes in transcript variants, which include genes for general metabolism. Furthermore, the expression of repressive epigenetic regulators increased with a concomitant decrease in gene activators that might be necessary to sustain the inherent plasticity of ESCs. Furthermore, we detected changes in microRNAs (miRNAs), with one set that targets early differentiation genes while another set targets pluripotency genes to maintain the unique ESC epigenotype. Such genetic and epigenetic events may contribute to a switch from a normal developmental program in adult cells during the formation of diseased tissues, including cancers.
Figures
Figure 1
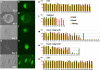
Morphology of ICM Outgrowth Bright field and fluorescence image of mouse E3.5 blastocyst (day 0 ICM outgrowth) (A and B), E4.5 blastocyst (C and D), day 3 ICM outgrowth (E and F), day 5 ICM outgrowth (G and H), and ESCs (I and J); real-time PCR measured gene expression in single cells of ICM outgrowth: Oct4, Sox2, and Nanog expression in 22 single E3.5 ICM cells (K), in 10 single E4.5 epiblast cells (L), in 26 single day 3 ICM outgrowth cells (M), in 17 single day 5 ICM outgrowth cells (N), and in 23 single ESCs (O). The y axis is normalized expression levels based on the mean expression values of all single cells for the given gene.
Figure 2
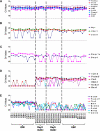
Gene Expression Measured by Real-Time PCR Gene expression measured by real-time PCR in single cells of fourteen ICM (E3.5), six day 3 ICM outgrowth cells, nine day 5 ICM outgrowth cells, and 14 ESCs (Table S1).
Figure 3
Transcriptome Analysis of ICM Outgrowth Cells (A) The principal component analysis of ICM outgrowth cells. The nine E3.5 ICM, three E4.5 Epiblast, two day 3 Oct4+Sox2+Nanog+ outgrowth cells, three day 5 Oct4+Sox2+Nanog+ outgrowth cells, two day 5 Oct4−Sox2−Nanog− outgrowth cells, and twelve ESCs are independently clustered (Table S2). (B) Expression dynamics of marker genes of early primordial germ cells (PGCs). Averaged expression of different individual cells was shown. The error bar represents the coefficient of variation (CV) between individual cells.
Figure 4
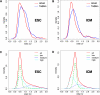
Plots of the Distribution of Coefficient of Variation between Individual Cells 158 (A) and 117 (B) genes are detected by TaqMan real-time PCR (Ct < 32 in at least half of the cells) in ESC and ICM cells, respectively, and used to compare cell-to-cell measurement variance. Density of coefficients of variation (CVs) for Ct measurements (blue) and RPM measurements representing RNA-Seq transcripts counts per million reads (red) across both ESC (A) and ICM (B) show a small difference between the two platforms. A similar representation is generated using the entire set of transcripts (24,435) separated into three categories: highly expressed genes (RPM > 10, aqua blue), mid-expressed genes (1 < PRM < 10, blue), and low-expressed genes (RPM < 1, green). CV density of each group of transcripts is represented for ESC (C) and ICM (D). The red curves are the sum of high-, mid- and low-expressed genes density of CVs. The mid-expressed genes tend to have higher cell-to-cell variations (Table S2). For each transcript, RPMs were used to calculate mean and standard deviation across cells of the same type. CV was defined as the ratio between the standard deviation and the mean value.
Figure 5
Splice-Specific Differential Expression The coverage plots of two junctions in (A) ESC, (B) E3.5 ICM, and (C) E4.5 epiblast of Dppa4 gene. The junction counts of Dppa4 transcript variant no. 1 (NM_001018002) are 12.47-fold more in ICM (88 reads, RPM = 150.16) than that in ESC (13 reads, RPM = 12.03), while the junction counts of Dppa4 transcript variant no. 2 (NM_028610) are 2.2-fold less in the ICM (35 reads, RPM = 59.72) than that in ESC (142 reads, RPM = 131.43, Table S2). RPM, read per million aligned reads.
Figure 6
Gene Network Analysis of Oct4 in Embryonic Stem Cell Pluripotency Pathway The 22 genes (including Oct4) up/downregulated for more than 4-fold when the pluripotent cells lose pluripotency (FC[day 5 Oct4+/day 5 Oct4−] > 4 or < 0.25, p < 0.01) were shown in red. The gray colored genes have FC[day 5 Oct4+/day 5 Oct4−] < 4 and FC[day 5 Oct4+/day 5 Oct4−] > 0.25 (Table S4). The p value was estimated using Ingenuity systems software (
). FC, fold change.
Figure 7
MicroRNA Expression in ICM and ESCs (A) miR-290 ∼−295 cluster microRNA expression in ICM and ESCs. (B) microRNAs showing significant differential expression between ICM and ESCs (p value < 0.01). The relative expression levels were shown (Table S5). The error bar represents the standard deviation calculated from three biological replicates.
Comment in
- Insightful tales from single embryonic cells.
Marks H, Veenstra GJ, Stunnenberg HG. Marks H, et al. Cell Stem Cell. 2010 May 7;6(5):397-8. doi: 10.1016/j.stem.2010.04.008. Cell Stem Cell. 2010. PMID: 20452308
Similar articles
- WNT/β-catenin signaling affects cell lineage and pluripotency-specific gene expression in bovine blastocysts: prospects for bovine embryonic stem cell derivation.
Madeja ZE, Hryniewicz K, Orsztynowicz M, Pawlak P, Perkowska A. Madeja ZE, et al. Stem Cells Dev. 2015 Oct 15;24(20):2437-54. doi: 10.1089/scd.2015.0053. Epub 2015 Aug 18. Stem Cells Dev. 2015. PMID: 26119137 - Rbm24 Regulates Alternative Splicing Switch in Embryonic Stem Cell Cardiac Lineage Differentiation.
Zhang T, Lin Y, Liu J, Zhang ZG, Fu W, Guo LY, Pan L, Kong X, Zhang MK, Lu YH, Huang ZR, Xie Q, Li WH, Xu XQ. Zhang T, et al. Stem Cells. 2016 Jul;34(7):1776-89. doi: 10.1002/stem.2366. Epub 2016 Mar 28. Stem Cells. 2016. PMID: 26990106 - Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells.
Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J. Babaie Y, et al. Stem Cells. 2007 Feb;25(2):500-10. doi: 10.1634/stemcells.2006-0426. Epub 2006 Oct 26. Stem Cells. 2007. PMID: 17068183 - Regulatory role of Klf5 in early mouse development and in embryonic stem cells.
Parisi S, Russo T. Parisi S, et al. Vitam Horm. 2011;87:381-97. doi: 10.1016/B978-0-12-386015-6.00037-8. Vitam Horm. 2011. PMID: 22127252 Review. - Genetic and epigenetic regulators of pluripotency.
Surani MA, Hayashi K, Hajkova P. Surani MA, et al. Cell. 2007 Feb 23;128(4):747-62. doi: 10.1016/j.cell.2007.02.010. Cell. 2007. PMID: 17320511 Review.
Cited by
- Measurement and modeling of signaling at the single-cell level.
Kolitz SE, Lauffenburger DA. Kolitz SE, et al. Biochemistry. 2012 Sep 25;51(38):7433-43. doi: 10.1021/bi300846p. Epub 2012 Sep 14. Biochemistry. 2012. PMID: 22954137 Free PMC article. Review. - Evaluation of commercially available RNA amplification kits for RNA sequencing using very low input amounts of total RNA.
Shanker S, Paulson A, Edenberg HJ, Peak A, Perera A, Alekseyev YO, Beckloff N, Bivens NJ, Donnelly R, Gillaspy AF, Grove D, Gu W, Jafari N, Kerley-Hamilton JS, Lyons RH, Tepper C, Nicolet CM. Shanker S, et al. J Biomol Tech. 2015 Apr;26(1):4-18. doi: 10.7171/jbt.15-2601-001. J Biomol Tech. 2015. PMID: 25649271 Free PMC article. - All models are wrong, but some are useful: Establishing standards for stem cell-based embryo models.
Posfai E, Lanner F, Mulas C, Leitch HG. Posfai E, et al. Stem Cell Reports. 2021 May 11;16(5):1117-1141. doi: 10.1016/j.stemcr.2021.03.019. Stem Cell Reports. 2021. PMID: 33979598 Free PMC article. Review. - Dynamic switching of active promoter and enhancer domains regulates Tet1 and Tet2 expression during cell state transitions between pluripotency and differentiation.
Sohni A, Bartoccetti M, Khoueiry R, Spans L, Vande Velde J, De Troyer L, Pulakanti K, Claessens F, Rao S, Koh KP. Sohni A, et al. Mol Cell Biol. 2015 Mar;35(6):1026-42. doi: 10.1128/MCB.01172-14. Epub 2015 Jan 12. Mol Cell Biol. 2015. PMID: 25582196 Free PMC article. - Novel Approaches to Profile Functional Long Noncoding RNAs Associated with Stem Cell Pluripotency.
Zhu Y, Yan Z, Tang Z, Li W. Zhu Y, et al. Curr Genomics. 2020 Jan;21(1):37-45. doi: 10.2174/1389202921666200210142840. Curr Genomics. 2020. PMID: 32655297 Free PMC article. Review.
References
- Aksoy I., Sakabedoyan C., Bourillot P.Y., Malashicheva A.B., Mancip J., Knoblauch K., Afanassieff M., Savatier P. Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases Pim-1 and Pim-3. Stem Cells. 2007;25:2996–3004. - PubMed
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. - PubMed
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases