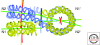Chromatin higher-order structure and dynamics - PubMed (original) (raw)
Review
Chromatin higher-order structure and dynamics
Christopher L Woodcock et al. Cold Spring Harb Perspect Biol. 2010 May.
Abstract
The primary role of the nucleus as an information storage, retrieval, and replication site requires the physical organization and compaction of meters of DNA. Although it has been clear for many years that nucleosomes constitute the first level of chromatin compaction, this contributes a relatively small fraction of the condensation needed to fit the typical genome into an interphase nucleus or set of metaphase chromosomes, indicating that there are additional "higher order" levels of chromatin condensation. Identifying these levels, their interrelationships, and the principles that govern their occurrence has been a challenging and much discussed problem. In this article, we focus on recent experimental advances and the emerging evidence indicating that structural plasticity and chromatin dynamics play dominant roles in genome organization. We also discuss novel approaches likely to yield important insights in the near future, and suggest research areas that merit further study.
Figures
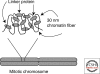
Figure 1.
The DNA path of a tetranucleosome as determined by X-ray diffraction. The structure consists of two stacks of nucleosomes, with linker DNA passing back and forth between them. Thus, the primary interactions occur between alternate rather than adjacent nucleosomes along the DNA strand, creating a zigzag architecture. From Schlach et al. 2005.
Figure 2.
EMANIC analysis of internucleosomal interactions. (A) Scheme of the EMANIC procedure. The two models for the structure of the chromatin 30-nm fiber, namely solenoid (Upper) and zigzag (Lower), lead to dominant i ± 1 and i ± 2 internucleosome interactions, respectively. (B–D) EM of nucleosome reconstitutes crosslinked in low salt without linker histone show few crosslinks. (E-I) With H1 present, ±2 interactions predominate. D′ and I′ diagram the nucleosome arrays corresponding to the adjacent EM images. From Grigoryev et al. 2009.
Figure 3.
Stereo pair of a section of nucleus in which a large region of chromosome is decorated with gold particles. Clear fiber-like structures of the order of 100 nm are seen. Scale is 500 nm. From Kireeva et al., 2008.
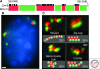
Figure 4.
(A) A large locus consisting of ∼4-Mbp region containing regions of gene “deserts” (red fluorescence) and gene clusters (green fluorescence) is seen in the nucleus in multiple configurations (C). In general, gene deserts are more closely associated with the heterochromatin at the nuclear periphery (B). Scale is 1 µm. From Shopland et al. 2006.
Figure 5.
Time sequence showing the extension of a metaphase chromosome in which the two ends are anchored by micropipettes. The diameter remains quite constant throughout. From Marko 2008.
Figure 6.
Extensible net model of mitotic chromosome structure derived from force-extension measurements. From Poirier and Marko 2002.
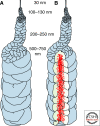
Figure 7.
Model of chromosome formation that incorporates the concept of a central axis enriched in condensins (red dots) and irregularly folded chromatin fibers. From Kireeva et al. 2004.
Figure 8.
The different forms of large-scale motion that contribute to chromatin dynamics. From Soutoglou and Misteli 2007.
Similar articles
- [Current insights into chromatin structure organization].
Ilatovskiĭ AV, Lebedev DV, Filatov MV, Petukhov MG, Isaev-Ivanov VV. Ilatovskiĭ AV, et al. Tsitologiia. 2012;54(4):298-306. Tsitologiia. 2012. PMID: 22724366 Review. Russian. - Three-dimensional genome organization in interphase and its relation to genome function.
Goetze S, Mateos-Langerak J, van Driel R. Goetze S, et al. Semin Cell Dev Biol. 2007 Oct;18(5):707-14. doi: 10.1016/j.semcdb.2007.08.007. Epub 2007 Aug 25. Semin Cell Dev Biol. 2007. PMID: 17905616 Review. - Chromatin Conformation Capture-Based Analysis of Nuclear Architecture.
Grob S, Grossniklaus U. Grob S, et al. Methods Mol Biol. 2017;1456:15-32. doi: 10.1007/978-1-4899-7708-3_2. Methods Mol Biol. 2017. PMID: 27770354 - Chromatin higher-order structures and gene regulation.
Li G, Reinberg D. Li G, et al. Curr Opin Genet Dev. 2011 Apr;21(2):175-86. doi: 10.1016/j.gde.2011.01.022. Epub 2011 Feb 20. Curr Opin Genet Dev. 2011. PMID: 21342762 Free PMC article. Review. - A physical model for the condensation and decondensation of eukaryotic chromosomes.
Mozziconacci J, Lavelle C, Barbi M, Lesne A, Victor JM. Mozziconacci J, et al. FEBS Lett. 2006 Jan 23;580(2):368-72. doi: 10.1016/j.febslet.2005.12.053. Epub 2005 Dec 27. FEBS Lett. 2006. PMID: 16387302
Cited by
- Insights into chromatin fibre structure by in vitro and in silico single-molecule stretching experiments.
Collepardo-Guevara R, Schlick T. Collepardo-Guevara R, et al. Biochem Soc Trans. 2013 Apr;41(2):494-500. doi: 10.1042/BST20120349. Biochem Soc Trans. 2013. PMID: 23514142 Free PMC article. Review. - How HP1 Post-Translational Modifications Regulate Heterochromatin Formation and Maintenance.
Sales-Gil R, Vagnarelli P. Sales-Gil R, et al. Cells. 2020 Jun 12;9(6):1460. doi: 10.3390/cells9061460. Cells. 2020. PMID: 32545538 Free PMC article. Review. - Regulation of nucleosome dynamics by histone modifications.
Zentner GE, Henikoff S. Zentner GE, et al. Nat Struct Mol Biol. 2013 Mar;20(3):259-66. doi: 10.1038/nsmb.2470. Nat Struct Mol Biol. 2013. PMID: 23463310 Review. - Chromatin Conformation in Development and Disease.
Boltsis I, Grosveld F, Giraud G, Kolovos P. Boltsis I, et al. Front Cell Dev Biol. 2021 Aug 4;9:723859. doi: 10.3389/fcell.2021.723859. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34422840 Free PMC article. Review. - Chromatin Ring Formation at Plant Centromeres.
Schubert V, Ruban A, Houben A. Schubert V, et al. Front Plant Sci. 2016 Feb 15;7:28. doi: 10.3389/fpls.2016.00028. eCollection 2016. Front Plant Sci. 2016. PMID: 26913037 Free PMC article.
References
- Belmont AS 2006. Mitotic chromosome structure and condensation. Curr Opin Cell Biol 18:632–638 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources